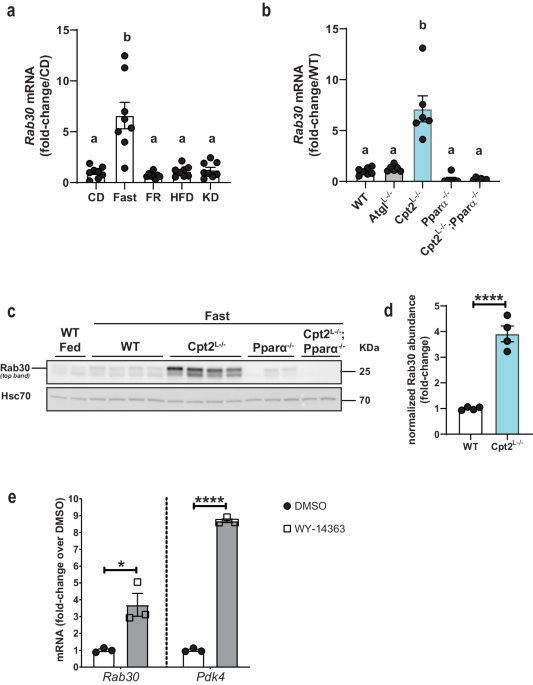
Rab30 is induced in the liver by fasting and requires Pparα
Previously, we showed that the liver specific loss of carnitine palmitoyltransferase 2 (Cpt2Lâ/â) and therefore hepatic mitochondrial long chain fatty acid β-oxidation resulted in dramatic fasting induced changes in the liver transcriptome and proteome18. In addition to changes in well characterized lipid metabolic genes, a relatively uncharacterized Rab GTPase, Rab30, was one of the most highly upregulated transcripts and proteins in Cpt2Lâ/â livers upon fasting (Supplementary Fig. 1a, b). To better characterize Rab30 regulation, we compared Rab30 expression across disparate metabolic conditions. First, our analysis of Rab30 expression in the livers of wildtype mice under different dietary states shows that Rab30 mRNA is induced specifically in the fasting state in the mouse liver as compared to chow-fed, re-feeding after a fast, high fat diet, and ketogenic diet (Fig. 1a)19. We next turned to mouse models of disrupted lipid metabolism and compared Rab30 expression in models of inhibited fatty acid oxidation with Cpt2Lâ/â mice and inhibited triglyceride hydrolysis with adipose triglyceride lipase liver-specific knockout (AtglLâ/â) animals. While both models exhibit a fatty liver following a fast20, only the loss of Cpt2 induced Rab30 above control levels (Fig. 1b), with fed state Rab30 mRNA levels similar between Cpt2Lâ/â and littermate controls (Supplementary Fig. 1c). These data show that Rab30 expression in the fasting state is exacerbated by a loss of fatty acid oxidation.
a Hepatic Rab30 mRNA from 20-week-old male C57Bl/6J mice under different dietary states. nâ=â8/group. CD control diet, Fast CD fed mice fasted overnight, FR CD fed mice fasted for 14âh then refed for 12âh; HFD high fat (60%) diet, KD ketogenic diet. Mice were placed on the respective diets for 12 weeks19. Values are meanâ±âSEM relative to CD. Letters indicate significance groups, where different letters represent statistical significance of pâ<â0.05 following analysis by Tamhaneâs T2 multiple comparisons test after Welch and BrownâForsythe ANOVA. b Hepatic Rab30 mRNA in fasted male mice (nâ=â7 for WT, nâ=â6 for all other genotypes). Values are meanâ±âSEM relative to WT fasted. Letters indicate significance groups by Tukeyâs multiple comparisons test following one-way ANOVA. WTâ=âCpt2 floxed. c Representative immunoblots of Rab30 expression in fed and 24âh fasted livers of male mice with Hsc70 as an equal protein loading control. WTâ=âRab30;Cpt2 floxed. d Quantitation of Rab30 band intensity of fasted WT and Cpt2Lâ/â samples (nâ=â4 males/genotype) in d, normalized to Hsc70 signal and represented as fold-change over WT signal. Values are meanâ±âSEM. Significance was determined by two-tailed unpaired t-test. ****pâ=â8.34âÃâ10â5. e Rab30 mRNA in primary mouse hepatocytes treated with DMSO vehicle control or the selective Pparα agonist WY-14643. Primary mouse hepatocytes (500,000 cells/one well of a 6-well plate) were treated with 10âμM WY-14643 or DMSO in Medium 199 supplemented with 10% FBS and 1% penicillin-streptomycin 100x solution for 16âh. Values are meanâ±âSEM relative to expression under control DMSO treatment. Significance was determined by two-tailed unpaired t-test comparing 3 wells of DMSO treated cells to 3 wells of WY-14643 treated cells for each gene. *pâ=â0.0167; ****pâ=â4.08âÃâ10â7. Pdk4 is a positive control for Pparα induction after WY-14643 administration. ANOVA tables and source data for relevant panels are provided as a Source Data file.
Compared to inhibition of triglyceride hydrolysis in AtglLâ/â, the loss of fatty acid oxidation by knockout of Cpt2 in the liver uniquely induces a Pparα transcriptional response20. As other groups have observed Rab30 to be among the top genes regulated by Pparα during fasting in the liver14,15,16,17, we examined the requirement of Pparα in the regulation of Rab30 in fatty acid oxidation deficiency in the livers of Cpt2Lâ/â;Pparαâ/â double-knockout mice (Fig. 1bâd). The loss of Pparα in a whole-body knockout (Pparαâ/â) failed to induce Rab30 in the liver following a fast and, furthermore, the loss of Pparα in Cpt2Lâ/â:Pparαâ/â double knockout mice ablated the Rab30 mRNA and protein induction by fasting. Finally, activating Pparα pharmacologically is sufficient to induce Rab30 mRNA in primary mouse hepatocytes (Fig. 1e). Taken together, these data show that Pparα is both required and sufficient to induce Rab30 in the liver following a fast.
Rab30 is localized to dynamic membranes from the Golgi apparatus
Rab30 has been reported to be a bona fide Golgi Rab21,22,23. Early dynamics studies in HeLa cells using photobleaching suggested that Rab30 is recruited to the Golgi from the cytosol but is not involved in vesicular or tubular exit from the Golgi21, while more recent studies indicate that it localized to vesicles through the endolysosomal pathway, including endosomes cycling the integral membrane protein TGN38 (human ortholog of mouse Tgoln1) between the Golgi and plasma membrane24, dynamic small intracellular vesicles marked by Tumor protein D52-like family members25,26,27, and even starvation and pathogen-induced autophagosomes28,29. BioID and other protein-protein interaction approaches in cultured human and Drosophila cells have also suggested Rab30 to interact with components of the exocyst and the Golgi-associated retrograde protein (GARP) complexes, indicating a potential for its involvement in the secretory and endocytic pathways30,31. Furthermore, Rab30 is observed to be localized to other membranes, such as to the apical pole in the Drosophila salivary gland, where it and other Rabs likely regulate Myosin V-dependent apical protein transport also in response to phosphatidylinositol levels28,32. The latter observation provides an interesting prospect of Rab30 as part of the machinery that maintains efficient vectorial transport and apical-to-basolateral polarity in mammalian epithelial cells such as hepatocytes. Therefore, given we understand how Rab30 expression is regulated in the mouse liver, we sought to define Rab30 localization, dynamics, and interactome in hepatocytes.
In order to better understand the localization of Rab30 in hepatocytes, we generated stably transfected AML12 cells, a cultured hepatic cell line, expressing an mScarletI-tagged Rab30. We observe that mScarletI-Rab30 localizes with the live Golgi stain C6-NBD-Ceramide in AML12 cells without perfect overlap (Fig. 2a). We next generated liver-specific adeno-associated viral vectors (AAV8) encoding mScarletI-tagged Rab30. Freshly isolated primary mouse hepatocytes expressing AAV-mScarletI-Rab30 demonstrated similar expression as AML12 cells in which mScarletI-Rab30 appears to be closely associating to but not overlapping with the Golgi stain (Fig. 2b, c). These observations are recapitulated in vivo with immunofluorescence staining of the cis-Golgi marker GM130 in the livers of fasted mice expressing AAV-mScarletI-Rab30, and we also find mScarletI signal to be polarized further out in the cytoplasm of hepatocytes (Fig. 2d).
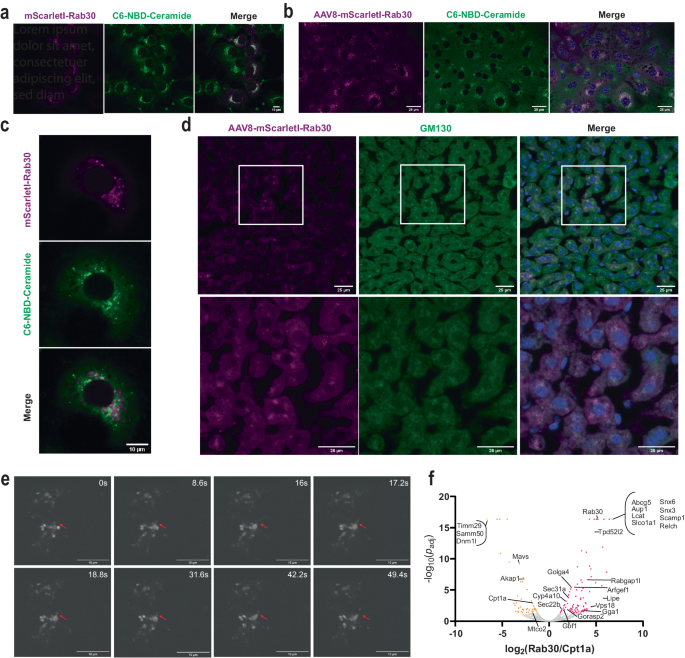
a Confocal microscopy image of AML12 cells stably expressing HA-mScarlet-Rab30 and stained with the live-cell Golgi marker C6-NBD-Ceramide. The experiment was repeated twice with cells in replicate wells. b Confocal microscopy image of mouse hepatocytes isolated from animals overexpressing with AAV8-mScarletI-Rab30 and stained with the live-cell Golgi marker C6-NBD-Ceramide and Hoechst 33342 nuclear stain. Experiment was performed once in replicate wells. c Zoomed-in image of a representative primary mouse hepatocyte overexpressing AAV8-mScarletI-Rab30 and stained with the live-cell Golgi marker C6-NBD-Ceramide from the experiment described in b. d Immunostaining of GM130 in 24âh fasted mouse liver tissue overexpressing AAV8-mScarletI-Rab30. Blue channel in the merge is Hoechst 33342 nuclear stain. The experiment was repeated in 2-3 sections from 2 different animals. e Still images from time lapse of primary mouse hepatocytes overexpressing AAV8-TBG-mScarletI-Rab30 following the progression of a putative mScarletI-Rab30-positive membrane protraction event (marked by red arrows) captured using the spinning disk confocal microscope CSU-W1 SoRa. 60x objective. The experiment was repeated twice with hepatocytes from 2 different animals. See also Supplementary Movies 1â3. f Volcano plot of proteins enriched in TurboID-Rab30 or -Cpt1a pulldowns. The bait proteins are highlighted in blue. Proteins detected exclusively in one pulldown are collapsed into one point marked in yellow. Significantly enriched proteins (padjâ<â0.05) are colored magenta for Rab30 and orange for Cpt1a. Statistical significance is reported as the adjusted p-value using the BenjaminiâHockberg correction for the false discovery rate (FDR). See Supplementary Data 2 for the list of proteins used to generate the plot. Source data are provided as a Source Data file.
To understand if Rab30-positive membranes are dynamically fusing to and exiting from the Golgi, we performed live-cell super-resolution imaging of mScarletI-Rab30 in primary mouse hepatocytes (Fig. 2e, Supplementary Movies 1â3). We captured dynamic budding of Rab30 vesicles and transient interactions with the Golgi: for example, an entire membrane protraction and vesiculation event of mScarletI-Rab30 membranes occurs in less than 50âs. An independent live-cell super-resolution experiment in which primary mouse hepatocytes expressing mScarletI-Rab30 and stained with a live cell Golgi marker provides stronger evidence in support of Rab30-marked membranes both dynamically interacting with and also budding from the C6-NBD-ceramide stained Golgi (Supplementary Movies 2, 3).
Given our observations of Rab30 distribution in hepatocytes, we wanted to identify what proteins interact with Rab30 and their intracellular localization. First, we performed a yeast two-hybrid screen of a constitutively-active Rab30 mutant (Q68L, amino acids 1â198 and lacking prenylation site) against a human liver cDNA library. We identified a small number of putative direct interactors including the Golgi coiled-coil membrane protein GOLGA5, consistent with a Golgi-specific function; somewhat unexpectedly, we detected albumin and the apolipoproteins B and A2, which we supposed would have been internal to a vesicle (Supplementary Data 1). Next, to identify the Rab30 interactome specifically in fasted state hepatocytes in vivo, we used the biotin-dependent proximity-labeling approach TurboID33. While we cannot differentiate between direct and indirect interactors by this technology, understanding what proteins are in the vicinity of Rab30 provides us with insight into potential processes that Rab30 may be involved in via association. To express the biotin ligase fusion protein in vivo, we generated liver specific AAV8s encoding TurboID-Rab30 and also TurboID-Cpt1a. We used TurboID-Cpt1a, which localizes to the outer mitochondrial membrane, as a compartmental control. We expressed each vector in 4 mice, then fasted the TurboID-Rab30 and -Cpt1a expressing mice for 21âh prior to injecting them with biotin and allowing a 3âh labeling period. At the end of the 3âh labeling period, we collected the livers, harvested total protein, and performed streptavidin pulldowns to enrich for biotinylated proteins. Western blot analysis from the livers of TurboID expressing animals and from streptavidin enrichment of biotinylated proteins shows both AAV expression and significant total biotinylation as compared to EGFP-injected animals (Supplementary Fig. 2a, b).
We used mass spectrometry to identify the proteins that bound to the streptavidin beads for each replicate. 278 proteins were significantly enriched in at least 2 replicates of TurboID-Rab30 samples, while 226 were enriched in at least 2 replicates TurboID-Cpt1a samples (Fig. 2f, Supplementary Data 2, Supplementary Data 3). The bait proteins Rab30 and Cpt1a were both significantly enriched in their respective pulldowns (Fig. 2f, Supplementary Fig. 2c). To identify the subcellular localization associated with potential interactors, the list of significantly enriched interactors for each TurboID fusion protein were submitted to the DAVID Bioinformatics Database34,35. As anticipated, we found that TurboID-Cpt1a interactors were significantly associated with the mitochondria, such as Samm50, Dmn1l, Timm29, Akap1, Mavs, and Mtco2 (Fig. 2f, Supplementary Fig. 2d). TurboID-Rab30 interactors, on the other hand, were more broadly associated with the secretory pathway, such as the ER, Golgi apparatus, endosome, vesicles, and cell junctions (Supplementary Fig. 2d).
Golgi-localized proteins and small GTPase effectors such as Golga4, Gorasp2, Rabgap1l, Gbf1, Arfgef1, and Gga1 were identified to be enriched in streptavidin pulldown samples for Rab30. Vesicle trafficking proteins such as Snx3, Snx6, Scamp1, and Relch were found exclusively in the streptavidin pulldowns for TurboID-Rab30 expressing animals, and Sec22b, Sec31a, Tpd52l2 (Tpd54), and Vps18 were also found to be significantly enriched. Interestingly, we also found proteins with functions in lipid and lipoprotein metabolism to be enriched as potential interactors for Rab30, such as such as the neutral sterol transporter localized to the bile canaliculus, Abcg5; the bile acid transporter localized to the basolateral membrane, Slco1a1; the hepatic secreted enzyme that esterifies the free cholesterol of lipoproteins, Lcat; a lipid droplet regulator and very low density lipoprotein assembly factor, Aup1; the triglyceride lipase, Lipe (Hsl); and the Pparα target gene Cyp4a10. Additionally, the Golgi-localized proteins Golga4 and Gorasp2 have been implicated in the regulation of lipoprotein metabolism previously36,37. Correspondingly, âlipid metabolic processâ and âfatty acid metabolic processâ are Gene Ontology terms associated with enriched interactors with Rab30 (Supplementary Fig. 2e). Together, these imaging and biochemical interaction data demonstrate that Rab30 is a dynamic interactor with the Golgi apparatus and also with proteins throughout the secretory pathway, likely localizing to cytoplasmic and post-Golgi vesicles in mouse hepatocytes. Furthermore, the interaction between Rab30 and known regulators of lipid metabolism supports the hypothesis for a role of Rab30 in contributing to hepatic lipid homeostasis. Further studies will be required to validate the location of the interactions and understand the functional role in hepatocyte biology. We now focus on the physiological implications of Rab30 on hepatic lipid metabolism.
Generation of Rab30 knockout mice
Given our observations that Rab30 (1) is a target of Pparα, a master transcriptional regulator of lipid catabolism and hepatocyte energy balance in the fasted state, (2) is dramatically upregulated in mice with deficient hepatic fatty acid oxidation, and (3) dynamically associates with the Golgi and proteins along the secretory pathway and regulators of lipid metabolism in live mouse hepatocytes, we hypothesized that loss of Rab30 in vivo might affect the secretion and/or turnover of lipids and proteins from the liver. Therefore, to determine the physiological role of Rab30 in response to fasting in the mouse liver, we generated Rab30 knockout (Rab30KO) and conditional knockout mouse (Rab30 ff) lines by CRISPR-Cas9 (Fig. 3a). Rab30KO mice were viable and fertile with an absence of Rab30 mRNA and protein (Fig. 3b, c). Mice containing the floxed Rab30 alleles were mated to mice expressing the albumin-Cre transgene to specifically knockout Rab30 in hepatocytes (Rab30Lâ/â) (Fig. 3d, e). Because we see a potentiated Pparα response in the livers of Cpt2Lâ/â fasted mice and because Rab30 expression depends on the activity of Pparα, we reasoned that we could use the genetic background of the Cpt2Lâ/â animals to amplify the effect of the loss of Rab30 in the liver. Therefore, Rab30 floxed mice were also mated to Cpt2 floxed animals carrying the albumin-Cre transgene to create liver-specific Rab30;Cpt2 double knockout (DKO) mice to determine the role of Rab30 induction in Cpt2Lâ/â mice (Fig. 3d, e). The loss of Rab30 in the germline, the liver, or in Cpt2Lâ/â liver had minimal effects on fed and fasting bodyweight, apart from Rab30KO males being larger on average than control males by 1.77âg in the fasted state (Supplementary Fig. 3a, c). Loss of Rab30 did not affect fed or fasting blood glucose measurements in males (Supplementary Fig. 3b); however, female blood glucose was increased in fasted Rab30KO female mice compared to littermates but decreased in DKO females compared to their controls (Supplementary Fig. 3d). While these data illuminate a potential sex difference in the response to fasting mediated by Rab30, we did not pursue further investigation of these discrepancies here because they are not related our main hypothesis regarding a role for Rab30 in the regulation of Pparα-dependent lipid and protein trafficking.
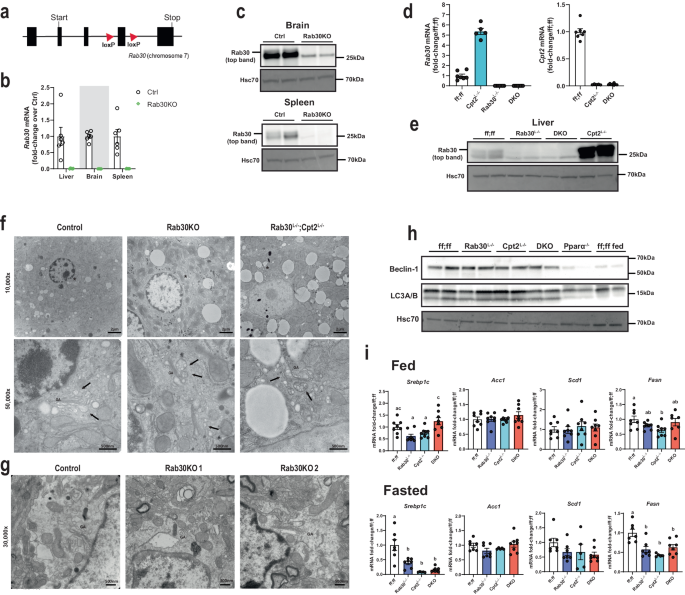
a Gene targeting strategy for the Rab30 gene, with loxP site insertion indicated by triangles. b Fasting Rab30 mRNA in the livers, brains, and spleens of whole-body Rab30 knockout male mice (Rab30KO) and littermates (Ctrl) (nâ=â6/genotype). Values are meanâ±âSEM relative to Ctrl for a given tissue. c Rab30 immunoblot in the brain and spleen of Rab30KO and control males. Hsc70 is an equal protein loading control. d Fasting hepatic qRT-PCR for Rab30 (left) and Cpt2 (right) mRNAs in male livers. Values are meanâ±âSEM relative to Rab30;Cpt2 floxed (ff;ff) animals. DKOâ=âRab30Lâ/â;Cpt2Lâ/â. nâ=â5 for Cpt2Lâ/â, nâ=â7 for ff;ff, and nâ=â8 for DKO. e Rab30 immunoblot in the livers of Rab30Lâ/â, Cpt2Lâ/â; DKO, and control (ff;ff) males. f Representative transmission electron micrographs of liver cells from 5â6-week-old 24âh fasted male knockouts and control males. Asterisks (*) in 10,000x denote region of 50,000x acquisition. GA, Golgi apparatus; arrows, vesicles. The experiment was performed in a total of 5 controls and 3 of each knockouts. g Representative transmission electron micrographs of hippocampi from 7-8-week-old control and 2 Rab30KO females under basal conditions taken at 30,000x. GA Golgi apparatus. The experiment was performed in a total of 3 females per genotype. h Western blot for autophagosome initiation markers in the livers of control fed male mice and fasted control (ff;ff), Rab30Lâ/â, Cpt2Lâ/â, Rab30;Cpt2 DKO, and Pparαâ/â male mice. Hsc70 is a protein loading control. i qRT-PCR of Srebp1c, Acc1, Scd1, and Fasn in fed and fasted livers of control (ff;ff), Rab30Lâ/â, Cpt2Lâ/â, and DKO males. nâ=â8/genotype for fed. nâ=â5 for Cpt2Lâ/â, nâ=â7 for ff;ff and Rab30Lâ/â, and nâ=â8 for DKO for fast. Values are meanâ±âSEM. Letters indicate significance groups by Tukeyâs multiple comparisons test following one-way ANOVA for each individual gene. ANOVA tables and source data for relevant panels are provided as a Source Data file.
The influence of Rab30 on Golgi-ultrastructure remains a point of contention throughout the literature. In cultured cells, siRNA knockdown of Rab30 revealed a Golgi-dispersion phenotype, but knockout of Rab30 by CRISPR/Cas9 in transformed cells showed no fragmentation of the Golgi21,24,38. To understand the contribution of Rab30 on Golgi structure in the mouse in vivo, we visualized the Golgi by transmission electron microscopy in livers of control, Rab30KO, and Rab30; Cpt2Lâ/â DKO mice and the hippocampus of control and Rab30KO mice (Fig. 3f, g). The Golgi apparatus appeared similar in both tissues between Rab30 knockout and wildtype mice. In conjunction with the data presented in Fig. 2, these data show that Rab30 associates with the Golgi but is not required for its ultrastructure in vivo.
Pparα regulates autophagosome formation in the mouse liver in response to fasting3,4,5. Given that the Golgi is also an important source for autophagic membrane lipids and proteins39, we asked if Rab30 was downstream of Pparα-directed autophagy. Fasting mRNA for autophagy related proteins, including Beclin-1 and LC3B, proteins responsible for the initiation of autophagosome formation, were found to be slightly increased (i.e., fold-change roughly <1.5) in both the Cpt2Lâ/â and DKO livers to similar extents, except for Lamp2, which was decreased in DKO livers (Supplementary Fig. 4). We next performed immunoblots against Beclin-1 and LC3A/B in wildtype fed livers and the fasted livers of wildtype, Rab30Lâ/â, Cpt2Lâ/â, Rab30;Cpt2 DKO, and Pparαâ/â animals (Fig. 3h). Beclin-1 and lipidated LC3 protein levels are not affected in fasted Rab30 knockout livers as compared to fed wildtype livers or fasted Pparα knockout livers, which exhibit decreased levels of both proteins. Taken together, our data does not support a role for Rab30 in Pparα-mediated autophagosome formation. However, as previously mentioned, Rab30 has been found to co-localize to autophagic membranes in HeLa cells28,29, and so it is possible that Rab30 could localize to the autophagosome following initiation and elongation or that the isolation membrane is scavenged from other organelles with the loss of Rab30. Another well-known role for the Golgi in regulating fatty acid metabolism is by affecting the processing of sterol regulatory element-binding protein 1 (Srebp1). Therefore, we determined the regulation of Srebp1 target genes in wildtype, Rab30Lâ/â, Cpt2Lâ/â, Rab30;Cpt2 DKO livers in the fed and fasted states (Fig. 3i). As Srebp1 target gene expression is largely unchanged with the loss of Rab30, we found no evidence to support a role of Rab30 in mediating Srebp1 processing.
Loss of Rab30 results in retention of secreted proteins
Next, we performed RNA-seq and proteomics on 24âh fasted livers from Rab30 knockouts, wildtype, and Cpt2Lâ/â animals for two main reasons. First, we were concerned that loss of Rab30 might influence the expression of other Rab proteins. Second, we predicted that if Rab30 was involved in intracellular membrane trafficking from the Golgi, then we would see a buildup of secreted proteins in the livers of Rab30 knockout mice.
Global analysis of the liver transcriptome by principle component analysis of normalized transcript levels revealed that the Rab30Lâ/â and Rab30KO clustered together with Rab30;Cpt2 floxed (ff;ff) and Rab30KO littermate controls (Rab30 Ctrl), while Cpt2Lâ/â and Rab30;Cpt2 DKO animals clustered together but apart from the single knockouts (Fig. 4a). These data are consistent with liver proteomic data that largely shows gene specific clustering (Fig. 4b). As we expected, these data suggest that loss of Rab30 does not drastically affect the fasted liver transcriptome.
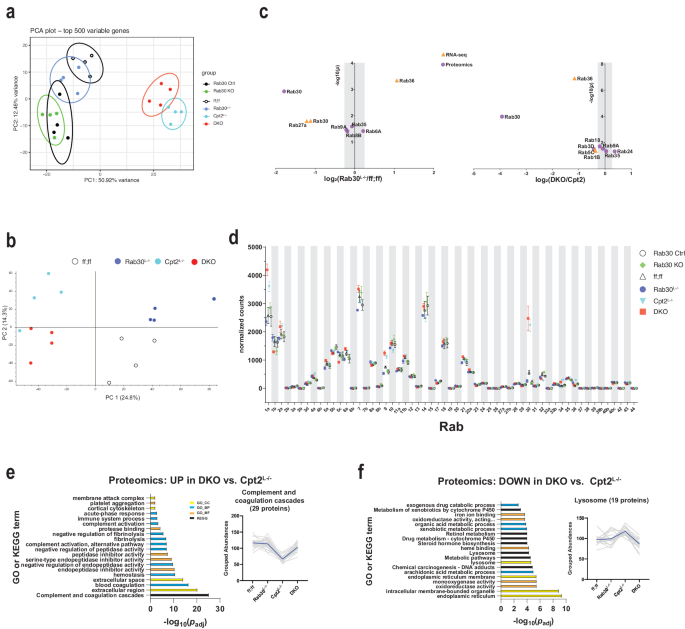
a Principal component analysis of RNA-seq on 24âh fasted livers from Rab30;Cpt2 floxed (ff;ff), Rab30KO littermate controls (Ctrl), Rab30Lâ/â, Cpt2Lâ/â, DKO, and Rab30KO male mice (nâ=â4/genotype). b Principal component analysis of proteomics on 24âh fasted livers from Rab30;Cpt2 floxed (ff;ff), Rab30Lâ/â (Rab30), Cpt2Lâ/â, and DKO mice (nâ=â4/genotype). c Volcano plot of significantly differentially expressed Rabs between Rab30Lâ/â and control (ff;ff) or DKO and Cpt2L-/â 24âh fasted livers in the RNA-seq (orange triangle) and proteomics (purple circle) datasets. Points outside of the gray bar represents a fold-change of at least 1.2. d Normalized read counts of Rab family members in RNA-seq of 24âh fasted livers. Values are the average of 4 samples/genotype ± SEM. Gene ontology and KEGG pathway analysis of proteomics comparing up- (e) and down- (f) regulated pathways in the DKO vs. Cpt2Lâ/â. Proteins with fold-change of at least 1.2 and pâ<â0.05 were submitted for pathway analysis to the DAVID functional annotation tool. Significantly enriched pathway terms are ranked against the Benjamini-adjusted p-value generated by the DAVID functional annotation tool. Top 20 terms by padj are presented. Colors represent pathway class as denoted in the legend. Normalized and scaled abundances vs. genotype plots depict the traces of all identified proteins within the given pathway, with the black line indicating the average grouped abundance of the proteins. For (e) mmu04610:Complement and coagulation cascades. For f GO:0005764~lysosome. p-values for pathway analysis and source data for relevant panels are provided in the Source Data file.
We directly investigated the transcriptomics and proteomics data set for differentially expressed Rab family members (Fig. 4c, d, Supplementary Fig. 5a). There were no significantly changed Rabs in the transcriptomics dataset when comparing Rab30KO livers to littermate control livers. When comparing Rab30Lâ/â to control (ff;ff) livers or DKO to Cpt2Lâ/â livers, we find a few changes in Rab mRNA and protein abundances; most were not greater than 1.2 fold-change (Fig. 4c, d, Supplementary Fig. 5a). In summary, loss of Rab30 does not appear to significantly alter the expression of other Rab family members in the liver. These data suggest that, at least at the expression level, the loss of Rab30 does not result in compensatory changes in other Rab family members; however, we do not know if loss of Rab30 affects their functions independently of expression level.
We then used the DAVID Bioinformatics Database34,35 to perform pathway analysis of up- and down-regulated proteins from our proteomic dataset. When comparing Rab30Lâ/â mice to control mice, we found no significantly enriched pathway terms upregulated in the liver knockouts. However, when comparing DKO mice to Cpt2Lâ/â mice, we find that there are pathway terms associated with hepatic secreted proteins upregulated in the fasted DKO livers (Fig. 4e). The GO cellular component term âGO:0005576~extracellular regionâ was the second-most enriched term by p-value and was comprised of 82 proteins. The KEGG term âmmu04610:Complement and coagulation cascadesâ was the most enriched by p-value and contained 29 proteins. This pathway is not significantly enriched on the transcriptomic level when comparing Cpt2Lâ/â to DKO (Supplementary Fig. 5b), indicating that the difference is not transcriptional in nature and possibly points to a defect in protein secretion and/or turnover. Furthermore, this term was also strongly downregulated on transcriptional levels in both the Cpt2Lâ/â and DKO fasted livers when compared to control animals; while this downregulation is upheld on the proteomic level for Cpt2Lâ/â mice, the loss of Rab30 in the DKO brings the protein abundances to about wildtype levels (Fig. 4e, trace of grouped abundances, and Supplementary Fig. 5c).
In terms of downregulated proteins, we find that proteins with pathway terms associated with the endoplasmic reticulum, intracellular membrane bounded organelle, and lysosome were significantly enriched in DKO animals compared to Cpt2Lâ/â (Fig. 4f). Specifically, the abundances of lysosomal resident or lysosomal-interacting proteins with the GO term âGO:0005764~lysosomeâ such as Lamp2, Syt11, Lipa, and Ctsf are decreased in DKO livers. Taken together, these data possibly indicate a disruption of trafficking from the Golgi with the loss of Rab30. We rationalize that we only observe this phenotype in the DKO livers because the demand for lipid and protein recycling for energetic substrates is exacerbated in hepatocytes that cannot generate ATP from lipids or rid the liver of excess fat through of β-oxidation, and perhaps there is increased demand of lysosomal degradation of proteins and lipid stores in the absence of Rab30-dependent membrane trafficking. In livers that are able to oxidize fats, the loss of Rab30 may be minimized due to increased metabolism or compensated by other mechanisms that maintain protein flux through the Golgi.
Rab30 influences circulating triglyceride and cholesterol
We next questioned if the defect in Golgi and endolysosomal trafficking of proteins could also relate to a role for Rab30 the mobilization of lipids in the liver in a Pparα-dependent manner. Therefore, we investigated if loss of Rab30 impacted lipid homeostasis in the liver and the serum of fasted mice. At the tissue level, the Cpt2Lâ/â and DKO livers are quantifiably larger and visibly paler than the control and Rab30Lâ/â livers (Fig. 5a, b). The basis for the dramatic increase in liver:body weight ratios after a fast in the Cpt2Lâ/â and DKO mice is predominantly due to fasting-induced hepatic lipid accumulation, as evidenced by the following rationale. First, there are no differences in liver:body weight ratios between knockouts and their controls in the fed state male mice (Supplementary Fig. 3e), and is consistent with previous observations described by our lab when initially characterizing Cpt2Lâ/â mice18. Next, examination of H&E stained sections of fed livers from control, Rab30Lâ/â, Cpt2Lâ/â, and DKO mice revealed no differences between genotypes (Supplementary Fig. 3f), while the fasting state histology showed that the Cpt2Lâ/â and DKO sections were comprised of cells that appeared to be swollen and lipid laden compared to the control and Rab30Lâ/â sections (Fig. 5c). BODIPY (Fig. 5c) and Oil Red O (Supplementary Fig. 3g) staining of fasted liver sections further indicate hepatic lipid accumulation in the DKO and Cpt2Lâ/â mice. The fasted state BODIPY 493/503 stained liver sections of the Rab30Lâ/â and control livers look overtly similar, while, as expected, both Cpt2Lâ/â and DKO livers contained cells bloated with lipids (Fig. 5c). Furthermore, as observed in BODIPY 493/503 staining of lipid droplets in fasted liver tissue (Fig. 5c), fasting induced significantly more lipid droplet accumulation in the liver with the loss of hepatic zonation in the periportal region in the Cpt2Lâ/â and DKO mice as compared to control or Rab30Lâ/â mice due to the imbalance of the inability to oxidize fatty acids with loss of Cpt2 and the influx of lipids. Oil Red O staining of the liver sections show similar results to the BODIPY stained liver sections (Supplementary Fig. 3g). Finally, while there is no difference in liver triglycerides between Rab30KO and Rab30Lâ/â and their respective controls, both the Cpt2Lâ/â and DKO livers exhibit increased hepatic triglycerides (Fig. 5d). Overall, as the hepatic triglyceride content is driven by Cpt2Lâ/â and not compounded by the loss of Rab30, we conclude that loss of Rab30 does not affect total hepatic triglyceride stores.
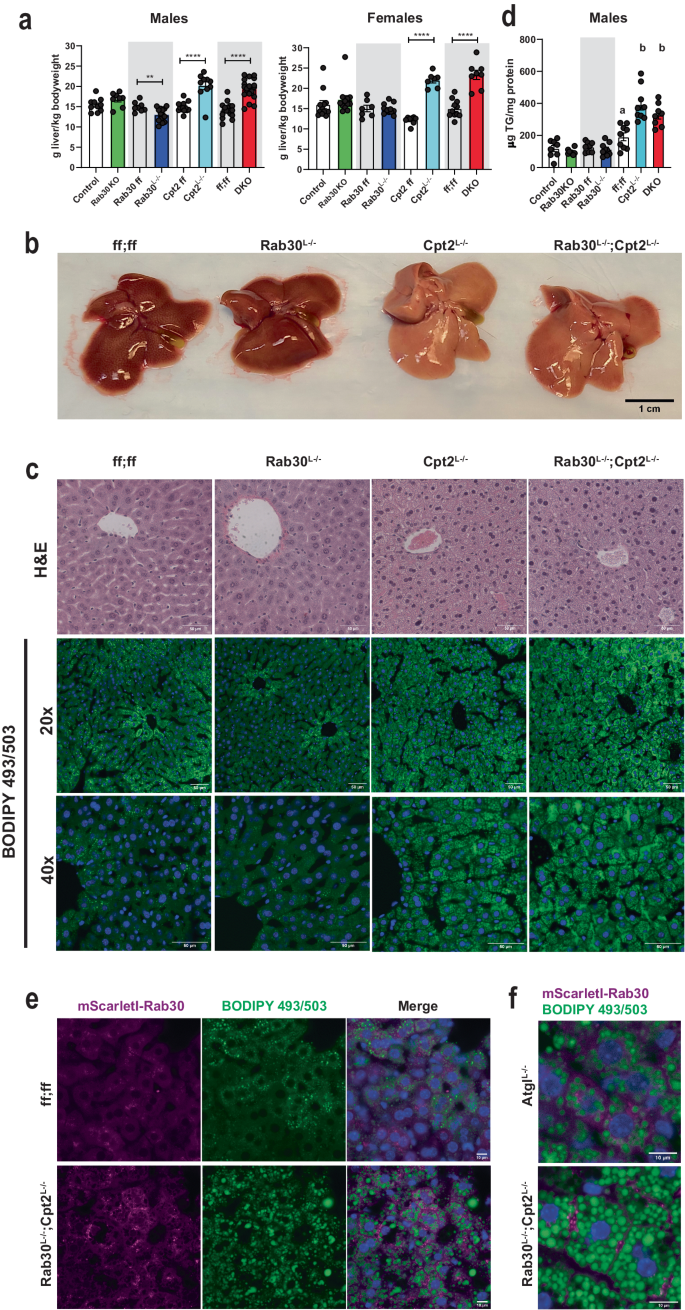
a Wet liver weights (large left lobe) normalized to body weight of fasted male and female mice. Data are represented as average ±SEM. Asterisks denote significance between knockouts and their littermate controls (Control, Rab30 ff, Cpt2 ff, and ff;ff for Rab30KO, Rab30Lâ/â, Cpt2Lâ/â, and DKO, respectively) by two-tailed t-test: *pâ<â0.05; **pâ<â0.01; ***pâ<â0.001, ****pâ<â0.0001. b 24âh fasted livers of control, Rab30Lâ/â, Cpt2Lâ/â, and Rab30;Cpt2 DKO female mice. c 24âh fasted histology of H&E and BODIPY 493/503 stained livers from control, Rab30KO, Cpt2Lâ/â, and Rab30;Cpt2 DKO male mice. Scale bar represents 50âµm. Blue in the BODIPY 493/503 images is Hoechst 33342 nuclear stain. Both experiments were performed in 2 mice per genotype. d Hepatic triglyceride levels in 24âh fasted male mice livers. Controls are Ctrl (nâ=â8) for Rab30KO (nâ=â8), ff (nâ=â8) for Rab30Lâ/â (nâ=â9), and ff;ff (nâ=â9) for Cpt2Lâ/â (nâ=â9) and DKO (nâ=â8) controls. Data are represented at average ± SEM. Significance was determined by two-tailed unpaired t-test for Ctrl vs Rab30KO (pâ=â0.32) and ff vs Rab30Lâ/â (pâ=â0.42). Significant differences between ff;ff, Cpt2Lâ/â, and DKO were determined by one-way ANOVA; letters indicate significance groups after Tukeyâs multiple comparisons test. e BODIPY 493/503 stained 24âh fasted livers from Rab30;Cpt2 floxed (ff;ff) and DKO females expressing mScarletI-Rab30 in hepatocytes by adenoassociated virus. Blue in merge is Hoechst 33342 nuclear stain. Scale bar represents 10âµm. Experiments were performed in 2 mice per genotype. f Comparison of mScarletI-Rab30 localization in 24âh fasted AtglLâ/â and DKO female livers stained with BODIPY 493/503. Experiment was performed in 1 AtglLâ/â female and 2 DKO females. If not reported in the legend, all n, p-values, ANOVA tables, and source data for relevant panels are provided in the Source Data file.
While Rab30 is clearly enriched near the Golgi in both cultured hepatocytes and wildtype fasted livers (Fig. 2aâd) and despite the fact that hepatic triglyceride content is not altered with the loss of Rab30, we questioned if Rab30 would be driven to localize with lipid droplets in livers lacking Cpt2 in a Pparα-dependent manner. We therefore expressed mScarletI-Rab30 by AAV8 in wildtype and DKO mice and stained their livers after a 24âh fast with BODIPY 493/503 to mark lipid droplets (Fig. 5e). As previously observed in Fig. 2d, Rab30 signal is observed to be polarized from the center of the cell throughout the cytoplasm in control fasted livers and does not co-localize with the modest accumulation of lipid droplets. In the lipid-laden DKO livers that exhibit potentiated Pparα transcriptional activity, Rab30 signal does not co-localize with lipid droplets as we initially wondered, but instead appears to be enriched at the cell periphery. This plasma-membrane localization is consistent with our TurboID-Rab30 data (Fig. 2), as we identified plasma membrane receptors and transporters to be interactors with Rab30 in the mouse liver. Interestingly, when we expressed mScarletI-Rab30 in AtglLâ/â livers, which also present a fasting-induced fatty liver but not Pparα induction, we do not find Rab30 to be enriched at the cell periphery (Fig. 5f).
While further experimentation is required to elucidate the mechanism behind the enrichment of Rab30 at the cell periphery only in mice lacking the ability to perform mitochondrial β-oxidation but not triglyceride hydrolysis, these results could imply an increased demand for lipid excursion in a Pparα-dependent manner in fasted livers that is exacerbated in the absence of β-oxidation and that Rab30 acts as part of this lipid disposal pathway. In fact, we have previously shown that Cpt2Lâ/â mice but not AtglLâ/â exhibit fasting-induced serum dyslipidemia20. We therefore asked if loss of Rab30 has an effect on circulating lipids. We quantified triglycerides, non-esterified fatty acids, cholesterol, and β-hydroxybutyrate in the serum from Rab30KO, Rab30Lâ/â, Cpt2Lâ/â, and DKO male mice compared to their controls. Rab30KO animals show a fasting-induced decrease serum triglycerides compared to littermates (Ctrl) (Fig. 6a). However, when comparing the Rab30Lâ/â to their controls (ff), there are no significant differences in the serum metabolites measured (Fig. 6b), indicating that the loss of Rab30 alone in the liver is not sufficient to influence fasting serum triglyceride levels, and that the decrease observed in the whole-body knockout is due to a cumulative effect of several contributing tissues. We next quantified these lipid species in Rab30;Cpt2 floxed (ff;ff), Cpt2Lâ/â, and DKO serum (Fig. 6c). Fasting ketone bodies were significantly decreased in the DKO and Cpt2Lâ/â as expected due to the suppression of ketogenesis in mice lacking hepatic Cpt2. As previously published, Cpt2Lâ/â mice have increased serum triglycerides and cholesterol levels18. Interestingly, the loss of Rab30 on the Cpt2Lâ/â background reverses the increased triglycerides and cholesterol levels back to control. Additionally, the DKO animals have increased NEFA compared to control and Cpt2Lâ/â. These differences are not observed in the fed state. By using the genetic background of Cpt2Lâ/â mice that exhibit a high induction of Rab30 upon fasting, we have discovered that hepatic Rab30 expression contributes to serum lipid levels during fasting.
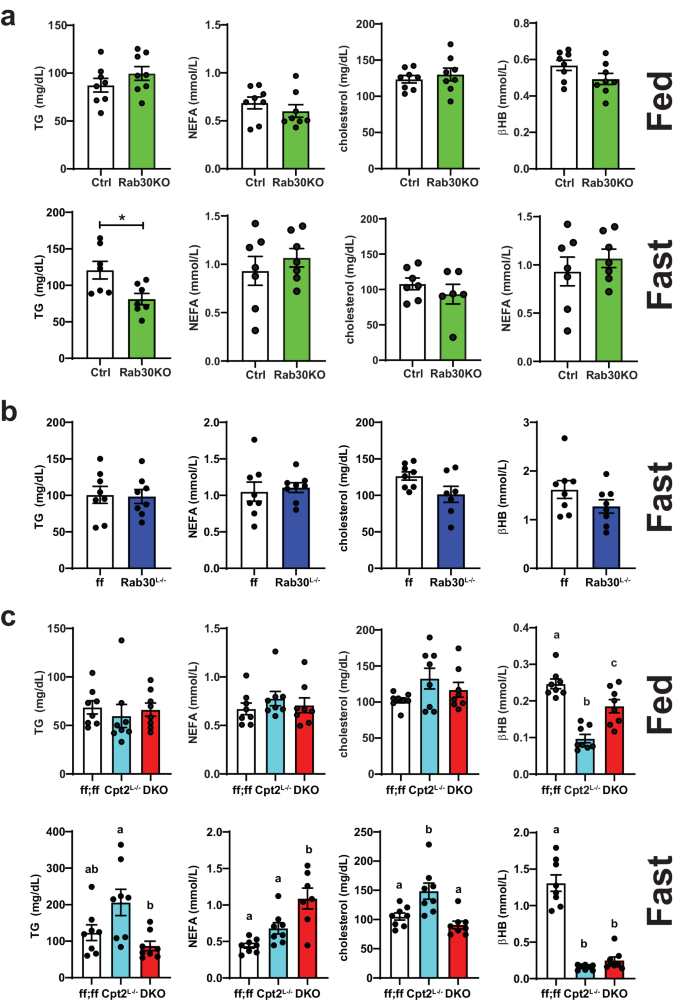
a Fed and fasting triglyceride (TG), nonesterified fatty acid (NEFA), cholesterol, and β-hydroxybutyrate (βHB) levels in the serum of male Rab30KO and littermate control (Ctrl) mice. Fed, nâ=â8/genotype. Fast, nâ=â7/genotype, except nâ=â6 Rab30KO for cholesterol. b Fasting TG, NEFA, cholesterol, and βHB levels in the serum of male Rab30Lâ/â and control (ff) mice. nâ=â8/genotype, except nâ=â7 Rab30Lâ/â for cholesterol. c Fed and fasting TG, NEFA, cholesterol, and βHB levels in the serum of male Cpt2Lâ/â, DKO, and control (ff;ff) mice. nâ=â8 for all genotypes across both states, except nâ=â7 DKO for cholesterol. In all panels, data are represented as average ±SEM. Significant differences (p-valueâ<â0.05) were determined by two-tailed unpaired t-test in a and b and by Tukeyâs multiple comparisonâs test following one-way ANOVA in c. *pâ<â0.05; shared letters indicate same significance level. p-values, ANOVA tables, and source data for relevant panels are provided in the Source Data file.
Loss of Rab30 impacts circulating ApoA4 abundance
As Rab30 does not localize to lipid droplets, it is not likely that Rab30 is influencing serum triglyceride levels through turnover of lipid droplets by direct interaction. Additionally, our proteomics data reveals that loss of Rab30 causes a retention of secreted proteins in the liver, indicating a potential transport defect. We therefore analyzed the serum proteome by separating serum-derived proteins on an SDS-PAGE gel and staining for total protein by Coomassie to see if we could identify secreted regulators of lipid metabolism influenced by loss of Rab30 (Fig. 7a). Overall, the banding pattern amongst genotypes appeared to be similar at the sensitivity afforded by this assay. However, we were intrigued to find that one ~45âkDa protein in particular was highly abundant in the Cpt2Lâ/â serum and appeared to be suppressed in the DKO serum (band âAâ), as well as further decreased in the control and Rab30Lâ/â animals. We excised this band, as well as the ~25âkDa protein labeled âBâ, which appears consistent amongst the genotypes. We identified these bands by mass spectrometry as ApoA4 (band âAâ) and ApoA1 (band âBâ). We then visualized ApoA4 levels in the serum of fasted control, Rab30Lâ/â, Cpt2Lâ/â, and DKO males and females by western blot (Fig. 7b, c, Supplementary Fig. 6aâd). The loss of Rab30 in the DKO suppresses the increase in the ApoA4 levels observed in the Cpt2Lâ/â serum.
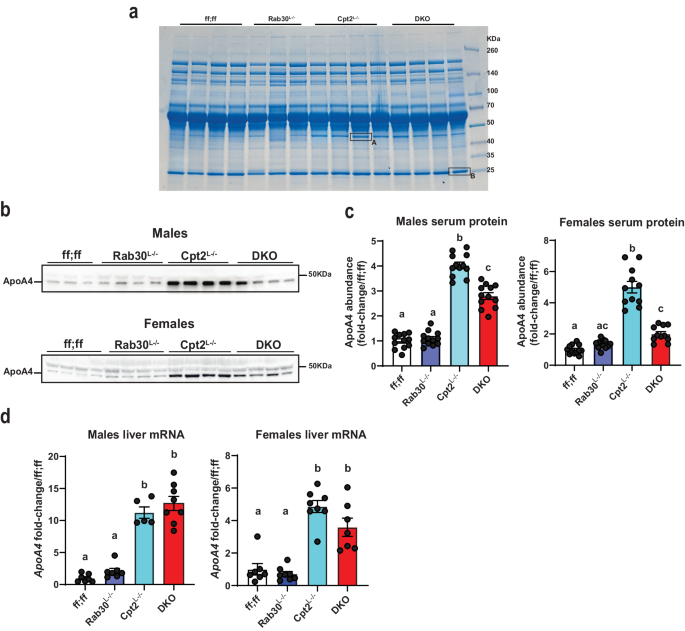
a Coomassie stained gel for total protein in the serum (0.5âμl/lane) of fasted male mice. nâ=â4 mice for ff;ff, Cpt2Lâ/â, and DKO, while nâ=â3 mice for Rab30Lâ/â. Bands A and B were excised for mass spectrometry analysis. b Representative ApoA4 immunoblot in the serum (1âμl/lane) of 24âh fasted ff;ff, Rab30Lâ/â, Cpt2Lâ/â, and DKO males and females. c Left, quantification of ApoA4 band intensity in serum of 24âh fasted males (nâ=â12 males/genotype); Right, quantification of ApoA4 band intensity in serum of 24âh fasted females (nâ=â12 females for ff;ff and Rab30Lâ/â, and nâ=â11 females for Cpt2Lâ/â and DKO). See Supplementary Fig. 6aâd for associated western blot for the quantitation. d Left, qRT-PCR of ApoA4 mRNA in 24âh fasted livers of males (nâ=â7 for ff;ff and Rab30Lâ/â, 5 for Cpt2Lâ/â, and 8 for DKO); Right, qRT-PCR of ApoA4 mRNA in 24âh fasted livers of females (nâ=â7 for ff;ff and DKO and 8 for Rab30Lâ/â and Cpt2Lâ/â). Letters indicate significance groups by Tukeyâs multiple comparisons test following one-way ANOVA. ANOVA tables and source data for relevant panels are provided as a Source Data file.
We investigated the expression of ApoA4 and other apolipoproteins in the livers of fasted mice. We performed qRT-PCR from fasted male livers for ApoE, ApoB, and a gene cluster of apolipoproteins conserved in mice and humans involving ApoA1, ApoC3, ApoA4, and ApoA5 (Fig. 7d, Supplementary Fig. 6e). ApoB, ApoA1, ApoC3, and ApoA5 mRNA varied little by genotype in males, while ApoE was suppressed ~40% in Cpt2Lâ/â and DKO livers. However, ApoA4 was the most highly regulated of the surveyed apolipoproteins and was significantly induced in the livers of both sexes of Cpt2Lâ/â and DKO animals (Fig. 7c, d). In agreement with the qRT-PCR data, transcriptomics data, and proteomics data, we observe the upregulation of hepatic ApoA4 in the Cpt2Lâ/â and DKO fasted male livers by western blot (Supplementary Fig. 6f, g). While ApoA4 is synthesized by the intestine and liver, hepatic expression of ApoA4 is hypothesized to decrease lipid burden of the liver by promoting excursion during fasting40,41. Therefore, these data indicate that ApoA4 could be playing a significant role in lipid turnover in the hepatocyte under the pathophysiological conditions of the Cpt2Lâ/â livers and its efficient trafficking requires Rab30 induction.