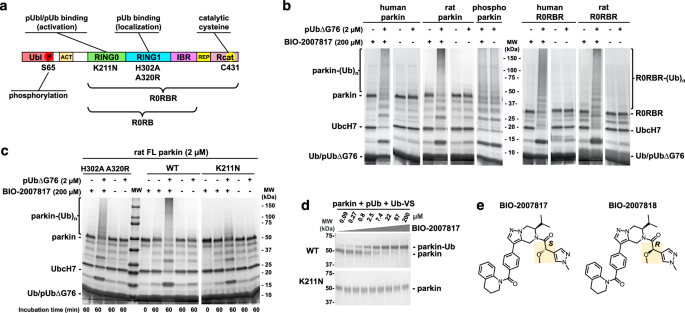
Ubl is not essential for parkin activation by BIO-2007817
Previous experiments showed that BIO-2007817 could trigger autoubiquitination activity of unphosphorylated parkin in the presence of pUb48. We confirmed those results with both human and rat parkin, which share 86% identity (Fig. 1b). At 200 µM, BIO-2007817 induced activity similar to phosphorylated (activated) parkin but the compound did not further stimulate the activity of phosphorylated parkin. As previously reported48, the activation was strictly dependent on the presence of pUb. To avoid charging of the E2 enzyme with pUb, a C-terminal deletion (pUbΔG76) was used. Activation could also be detected in ubiquitin-vinyl sulfone (Ub-VS) assays51 that measure the accessibility of Rcat and are not dependent on the upstream components of the ubiquitination cascade (Fig. 1d). We next asked if the parkin Ubl domain was required for activation. We observed equal autoubiquitination activity of full-length and R0RBR parkin (lacking the Ubl domain) in both the human and rat proteins (Fig. 1b). Thus, the Ubl domain is not implicated in the mechanism of activation.
BIO-2007817 requires pUb bound to RING0 domain
Phospho-ubiquitin can bind two distinct sites in parkin. The high-affinity site on RING1 mediates the recruitment of parkin to the mitochondria19. With an affinity of around 20 nM, it is formed by residues H302 and R305 and promotes detachment and phosphorylation of the Ubl domain52. The second site is located on the RING0 domain and regulates parkin activity. While the second site typically binds pUbl (when parkin is phosphorylated), we previously demonstrated that it can also bind pUb49. The site is formed by residues K211, R163 and K161 and with an affinity of around 400 nM for pUb and 1 µM for pUbl49,50. This binding forces the dissociation of Rcat domain and the release of parkin ligase activity.
We investigated which of the two sites is needed to promote activation by comparing the effects of a RING1 double mutation (H302A A320R) and a RING0 mutation (K211N). In the absence of compound, none of the parkin variants—wild-type, H302A A320R, or K211N—can autoubiquitinate in the presence of pUb (Fig. 1c). Upon addition of BIO-2007817, the double mutant H302A A320R was activated to close to wild-type levels as judged by the disappearance of unmodified parkin and appearance of a high molecular weight smear of autoubiquitinated parkin. In contrast, the K211N parkin mutant was strongly impaired with an absence of high molecular weight parkin adducts (Fig. 1c). Thus, pUb binding to RING0 but not RING1 is required for activation by BIO-2007817. We confirmed this result in the Ub-VS assay that measures Rcat release. At 2.5 µM BIO-2007817, half of wild-type parkin was modified by the Ub-VS reagent while the K211N mutation completely prevented parkin modification even at 200 µM (Fig. 1d). Taken together, these results demonstrate that activation by BIO-2007817 requires the binding of an exogenous pUb molecule to the RING0 binding site of parkin.
BIO-2007817 acts a molecular glue for RING0 and pUb/pUbl
We measured the affinity of BIO-2007817 for parkin using isothermal titration calorimetry (ITC) experiments with a parkin construct R0RB that lacks both the Ubl domain and the Rcat domain (Fig. 2, Supplementary Table 1). Since pUb and Rcat compete for binding to RING0, deleting the Rcat domain favors pUb binding. The initial experiment measured BIO-2007817 binding to R0RB with pUb bound to both the RING1 and RING0 sites (R0RB + two pUb). BIO-2007817 bound the complex with 10 nM affinity and one-to-one stoichiometry (Fig. 2a). The interaction was highly specific as the inactive diastereomer BIO-200781848 (Fig. 1e) bound with 150-fold less affinity (Fig. 2a).
Isothermal titration calorimetry measurements with different parkin complexes and titrants (in italics). a BIO-2007817 binds to the R0RB:2×pUb complex with 150-fold higher affinity than the diastereomer BIO-2007818. b High-affinity binding requires the presence of pUb. c The K211N mutation in the RING0 pUb/pUbl-binding site prevents high-affinity binding. d BIO-2007817 also binds to the R0RB:pUbl complex. e Inclusion of ACT element with pUbl decreases the affinity of BIO-2007817 10-fold. f The F146Y mutation in RING0 ACT-binding site decreases the affinity 80-fold. g pUbl binds weakly to R0RB49,50. h Inclusion of BIO-2007817 increases the affinity of pUbl binding by 2400-fold. Protein concentrations and binding parameters are given in Supplementary Table 1.
We next asked if pUb was required for BIO-2007817 binding (Fig. 2b). In the absence of pUb, the affinity was decreased over 5000-fold (to the limit of detection), mirroring the requirement for pUb observed in autoubiquitination experiments. Loss of the RING0 pUb-binding site (due to the K211N mutation) similarly prevented binding (Fig. 2c). Although BIO-2007817 had no effect on phosphorylated parkin, surprisingly, the phosphorylated Ubl domain (pUbl) added in trans could replace pUb. In ITC experiments, BIO-2007817 bound to the R0RB:pUbl complex with the same affinity as the pUb complex (Fig. 2d). As the R0RB fragment does not undergo any conformational rearrangements and pUbl binds only to RING0, this suggests that BIO-2007817 binds at the RING0/pUb (or RING0/pUbl) interface.
Additional insight into the site of BIO-2007817 binding came from the observation that inclusion of the ACT region with the pUbl domain (pUbl-ACT) decreased the affinity of BIO-2007817 binding 10-fold (Fig. 2e). In contrast, in the absence of compound, the ACT improves the affinity of pUbl binding to R0BR three-fold50. This suggests they compete for a common binding site. Identification of the binding site was confirmed by a mutation in ACT-binding site. Addition of a single oxygen atom (F146Y) decreased BIO-2007817 binding 10-fold (Fig. 2f). As F146 is surface-exposed and does not make contacts with pUb or pUbl in crystal structures of parkin, this strongly suggests that F146 directly binds BIO-2007817.
To directly test if BIO-2007817 behaves as a molecular glue, we measured the affinity of the pUbl domain binding to RING0 in the R0RB construct. As previously reported49,50, pUbl in trans has low micromolar affinity (Fig. 2g). This contrasts with the situation in intact phosphorylated parkin when the pUbl domain is present in cis. Inclusion of a saturating concentration (200 µM) of BIO-2007817 increased the pUbl binding affinity by more than three orders of magnitude, demonstrating that the compound acts as a molecular glue (Fig. 2h).
Mechanism of activation by THPP compounds
Hydrogen-deuterium exchange mass spectrometry (HDX-MS) has been instrumental for characterizing the conformational changes during parkin activation29,30,31. HDX-MS measures solvent exposure which provides information on protein structure and interdomain contacts. We performed a series of HDX-MS experiments to resolve the mechanism of parkin activation by THPP compounds (Fig. 3). As previously observed, addition of pUb to parkin increased deuteration of the Ubl domain (Fig. 3a). This reflects the release of the Ubl from its binding site on RING1, which facilitates its phosphorylation by PINK129,30,31. There was no increase in deuteration of Rcat domain as pUb binding to RING1 is not sufficient to activate parkin. In contrast, addition of BIO-2007817 in the presence of pUb, increased deuteration of the catalytic Rcat domain, reflecting its release from RING0 (Fig. 3b). The effect of BIO-2007817 was strictly dependent on the presence of pUb; no changes in parkin deuteration were observed upon addition of BIO-2007817 alone (Fig. 3c). The increase in Rcat exposure was also completely blocked by the K211N mutation that prevents pUb binding to RING0 (Fig. 3d) or by substitution of BIO-2007818 with the inactive diastereomer (Fig. 3e). Thus, as observed in the autoubiquitination assays (Fig. 1), the combined action of pUb and BIO-2007817 is required to activate parkin. This occurs through the release of the Rcat domain from its site on RING0 in a manner analogous to activation by parkin phosphorylation29,30,31 (Fig. 3f).
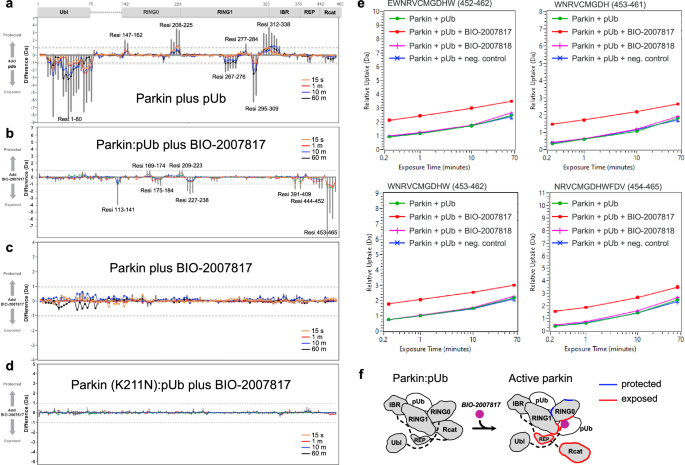
a Changes in solvent exposure of the parkin complex upon addition of pUb at four time points (15 s, 1 min, 10 min, 60 min) and the sum of the four time points (gray bars). The largest changes occur in the Ubl domain, which is released by pUb binding, and RING1, which is the site of pUb binding. b Changes in solvent exposure upon addition of BIO-2007817 in the presence of pUb. The largest changes occur in the Rcat domain. c, d Addition of BIO-2007817 does not change solvent exposure in the absence of pUb or in the parkin K211N mutant. e HDX-MS analysis of individual peptides from the parkin Rcat domain show solvent exposure in the presence of BIO-2007817 but not the inactive diastereomer BIO-2007818. f Mapping of increased (red) and decreased (blue) exchange upon BIO-2007817 addition to parkin:pUb (b).
Crystal structure of parkin R0RB:2×pUb complex with BIO-1975900
We turned to crystallography to understand how BIO-2007817 acts as a molecular glue. As crystallization trials with BIO-2007817 were unsuccessful, we expanded the screen to include related compounds with different ethanone substituents. We obtained crystals with the compound BIO-1975900 bound to rat parkin R0RB with two pUb and solved the structure by molecular replacement (Fig. 4; Supplementary Table 2). BIO-1975900 is composed of a central tetrahydropyrazolo-pyrazine core, decorated by a tetrahydroquinoline head and a benzyl and ethanone tail (Fig. 4a). It has roughly 20-fold lower affinity with an EC50 of 2.9 µM in in vitro TR-FRET activity assays compared to 0.15 µM for BIO-200781748.
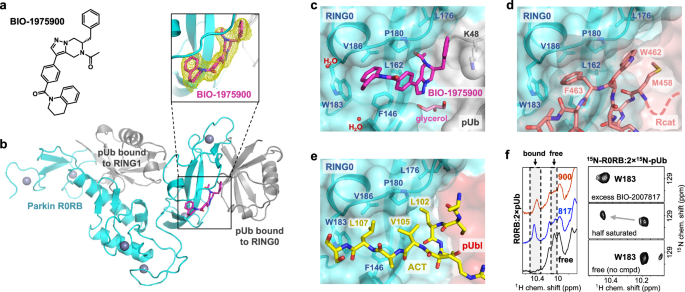
a Chemical structure of BIO-1975900. b Crystal structure of rat R0RB:2×pUb with BIO-1975900. Inset shows the 2.5 Å omit map of BIO-1975900 contoured at 3 σ. c Details of the BIO-1975900 (magenta) bound to a hydrophobic groove formed by residues from RING0 and a lysine from pUb. A glycerol and two water molecules were observed at the binding site. d Overlay of the inactive parkin structure (PDB 5N2W)26. The C-terminus of the Rcat domain (salmon) occupies the BIO-1975900 binding site. e Overlay of the active, phosphorylated parkin:pUb structure (PDB 6GLC). The ACT element (yellow) binds the same site as BIO-1975900. f NMR spectra of THPP binding. 1D NMR spectra (left panel) of the downfield region upon addition of BIO-1975900 or BIO-2007817. 2D 1H-15N correlation NMR spectra (right panel) of the tryptophan W183 indole amide titrated with BIO-2007817.
Overlay of the electron density of the unliganded structure50 (PDB 7US1) showed additional electron density at the RING0-pUb interface corresponding to BIO-1975900 (Fig. 4b). The compound binds a hydrophobic groove composed of three regions: a RING0 W183 pocket, a P180 pocket, and a pUb K48 pocket (Fig. 4c). The tetrahydroquinoline group contacts F146, P180, W183, and V186. Replacement of F146 by tyrosine introduces a polar group and disrupts binding as observed in ITC experiments. The ethanone group inserts into a hydrophobic pocket formed by parkin RING0 residues L162, V164, L176, P180, and F208. The benzyl moiety extends to sit in a pocket sandwiched between the L176 residues of RING0 and residue K48 of the pUb molecule bound to RING0 (Fig. 4c). In the difference density, we also identified a glycerol molecule from the cryoprotectant bound near the THPP central ring. The glycerol bridges the backbone amide of L162 and the guanidino group of R163 in the RING0 phosphoserine binding site.
We used NMR spectroscopy to confirm that BIO-1975900 and BIO-2007817 bind at the same location. The proton NMR spectrum shows similar chemical shift changes in the downfield region upon addition of the two compounds (Fig. 4f, left panel). 2D NMR with 15N-labeled R0RB:2×pUb protein confirmed that the shifting signal was from W183, which is the only tryptophan present in R0RB:2×pUb (Fig. 4f, right panel). Addition of half an equivalent of BIO-2007817 to 15N-labeled R0RB:2×pUb split the W183 side chain signal at 10.2 ppm into two peaks, indicative of slow-exchange kinetics typical for nanomolar binding affinity. Addition of excess of BIO-2007817 shifted the signal to 10.5 ppm, as observed in the 1D spectra (Fig. 4f, left panel). The downfield chemical shift change arises from the interaction between the tetrahydroquinoline moiety of either BIO-2007817 or BIO-1975900 and the W183 side chain. This confirms that BIO-2007817 binds at the RING0/pUb interface in the same manner as BIO-1975900.
The crystal structure highlights the importance of the hydrophobic groove that binds BIO-1975900 for controlling parkin activity. In autoinhibited structures of parkin, Rcat residues bind the groove: W462 forms hydrophobic interactions with RING0 while F463 contacts RING0 W183 (Fig. 4d). The F463 is positioned almost identically to the tetrahydroquinoline group of BIO-1975900. BIO-2007817 and Rcat compete for binding the same site on RING0.
The structure also explains the competition observed in ITC experiments between ACT and BIO-2007817 binding. The ACT, formed by residues 102–109 in the Ubl-R0RBR linker, binds to the same hydrophobic groove than BIO-1975900 (Fig. 4e), as observed in the active, phosphorylated parkin:pUb structure (PDB 6GLC). ACT L107 contacts W183, L102 contacts F208, and V105 stacks against RING0 P180. The molecular mimicry between the compound and the hydrophobic residues of the ACT element explains, in part, why the compound is unable to increase the activity of phosphorylated parkin.
The importance of the hydrophobic groove in parkin activation has been observed before. The mutation F146A weakens the hydrophobic interaction with Rcat F463 and, consequently, promotes parkin activation16. Cells transfected with the F146A mutant showed increased recruitment of parkin to mitochondria and mitophagy36. The F146A mutation also rescues mitochondrial recruitment and mitophagy in parkin mutants with defective Ubl (R42P, V56E, S65A, and ∆Ubl) or RING0 pUbl/pUb-binding sites (K161N and K211N)35,36.
Structure-activity relationship of THPP compounds
We used molecular docking to explore the structure-activity relationship of related THPP compounds. Parkin activity was previously determined for compounds from two subseries: BIO-2007817-related subseries 1, which has an isopropyl group attached to its pyrazolo-pyrazine moiety, and BIO-1975900-related subseries 2, in which the isopropyl is replaced by a benzyl group46. Additional compounds from both subseries were selected to rationalize how chemical modifications in the three interaction regions affect their activity (Fig. 5a, Supplementary Fig. 1). To validate the protocol, BIO-1975900 was first docked into the R0RB:2×pUb crystal structure using the standard precision ligand docking module in Glide. The docked compound had favorable energies (Fig. 5c) and superimposed well (less than 0.3 Å deviation) with the experimentally determined structure.
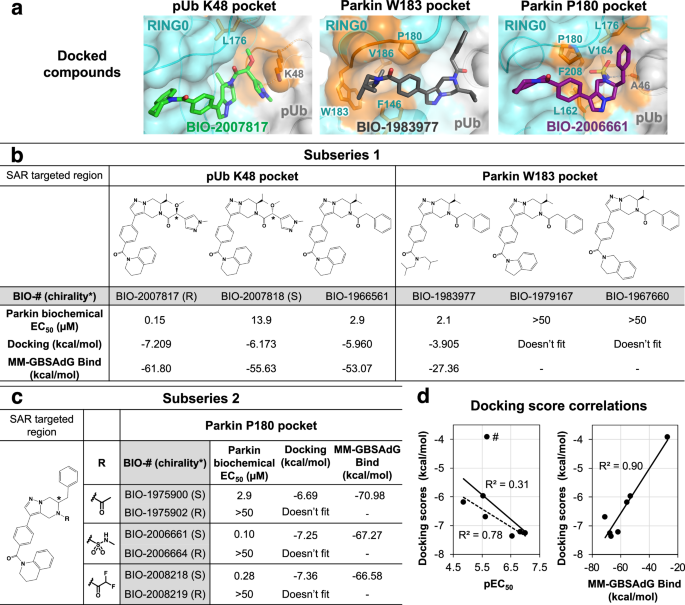
a Docking-based positioning of BIO-1975900 related-compounds. Comparison of biochemical potency (EC50 in parkin autoubiquitination assays) and the docking scores for subseries 1 (b) and subseries 2 (c) compounds. Autoubiquitination activity profiles are shown in Supplementary Fig. 1. d Correlation between docking scores, biochemical potency, and MM-GBSA scores. BIO-1983977 lacks the THPP group and is an outlier: removing it (marked #) improves the correlation between the docking score and pEC50 (dashed line).
Subseries 1-compound BIO-2007817 was docked into the prepared R0RB:2×pUb protein grid (Fig. 5a left panel, Supplementary Fig. 2). NMR experiments with BIO-2007817 revealed hindered rotation with the methoxy-methylpyrazine-ethanone moiety adopting cis and trans conformations (Supplementary Fig. 3). In the docked structure, BIO-2007817 is in the cis conformation, suggesting a conformationally locked compound could likely improve potency further. The isopropyl moiety points towards parkin V164 and L176 and the methoxy-methylpyrazine-ethanone groups are adjacent to the positively charged side chain of pUb K48, forming a cation-pi interaction. The pi-stacking interactions between tetrahydroquinoline group and W183 and F146 are similar to BIO-1975900. In the weakly binding BIO-2007818 diastereomer, the oxygen of the methoxy group points away from pUb K48. The loss of the electrostatic interaction is reflected in lower docking and MM-GBSA scores compared to BIO-2007817 (Fig. 5b). This agrees with the limited effect of BIO-2007818 on parkin activation48 and its reduced binding in ITC experiments (Fig. 2a). Replacement of the methoxy and methyl-pyrazolyl groups with a phenyl moiety (BIO-1966561) also decreased the docking score and EC50 (Fig. 5b).
Docking of BIO-1983977 (Fig. 5a, middle panel) demonstrates that the hydrophobic diisobutylamine functional group fits in the W183 pocket. The predicted binding is weaker than compound BIO-1966561 (identical except for the tetrahydroquinoline head group) but BIO-1983977 was similar in biochemical assays (Fig. 5b). The inactive compounds BIO-1967660 and BIO-1979167 could not be docked due to steric clashes with the sides of the W183 pocket.
Modulation of compound interactions with the parkin P180 pocket was investigated by docking subseries 2 compounds which are most closely related to the crystallized ligand BIO-1975900 (Fig. 5c). In contrast to the ethanone group of BIO-1975900, which showed limited interactions with the P180 pocket (Fig. 4c), the methylsulfonamide moiety of BIO-2006661 formed a hydrogen bond with the backbone carbonyl of pUb A46 (Fig. 5a, right panel). The additional interaction would explain the higher potency of BIO-2006661 relative to BIO-1975900 (EC50 of 0.1 µM and 2.9 µM). BIO-2008218, which contains a difluoro-ethanone moiety, showed a similar increase in potency. Positioning of the subseries 2 benzyl group was critical for binding. BIO-1975902, BIO-2006664, and BIO-2008219, R-enantiomers of active S-compounds, were inactive in biochemical assays and could not be docked. In general, the calculated binding interactions correlated well with the biochemical EC50, suggesting that docking could be used for shape-based screening of new parkin activators (Fig. 5d).
BIO-2007817 rescues ubiquitination and mitophagy by parkin Ubl mutants
The observation that BIO-2007817 can activate parkin in the presence of pUb suggested that it might rescue the activity of parkin variants without a functional Ubl domain. We tested this hypothesis using an in organello assay that measures parkin ubiquitination of the mitochondrial protein mitofusin-2 (Mfn2)36. As substrate ubiquitination and autoubiquitination rates can differ, it was important to test the activity of the compounds on a physiologically relevant substrate. In the assay, HeLa cells are stimulated by treatment with mitochondria depolarizer CCCP and then the mitochondria are isolated and added ex vivo to a ubiquitination reaction mix including parkin with or without activator compounds (Fig. 6a).
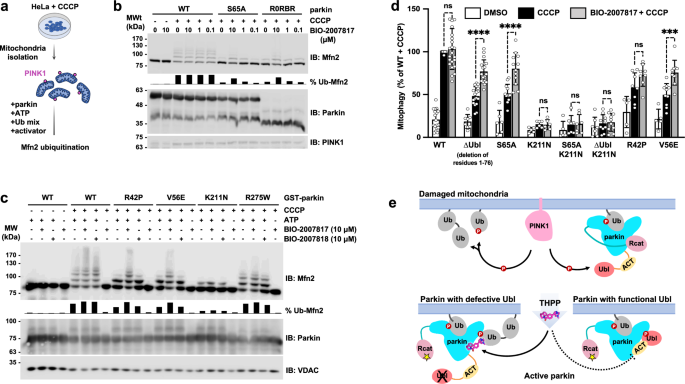
a Schematic of in organello ubiquitination assay of parkin. PINK1 expression is induced in cultured cells by CCCP and parkin activity measured by ubiquitination of mitofusin-2 (Mfn2). b Immunoblots of in organello assays showing the rescue of ubiquitination activity of the non-phosphorylatable parkin mutants, S65A and R0RBR, by BIO-2007817. Quantification of the percentage of ubiquitinated Mfn2 is shown as chart bar. c Immunoblots of in organello assays showing the rescue of ubiquitination activity of Parkinson’s disease parkin mutants, R42P and V56E, by BIO-2007817, but not by the diastereomer, BIO-2007818. Quantification of the percentage of ubiquitinated Mfn2 is shown as chart bar. d Quantification of average percentage of mitophagy detected by fluorescence‐activated cell sorting (FACS) of mitochondrially targeted mitoKeima U2OS cells expressing transient WT or mutant GFP‐parkin. Cells were treated with DMSO (white), CCCP (black), or pre-treated with BIO-2007817 followed by CCCP treatment (gray). Percentages were normalized to WT Parkin pre-treated with DMSO followed by CCCP treatment. A partial rescue by BIO-2007817 was observed in cells expressing parkin with defective Ubl. Error bars indicate s.e.m. For statistical analysis, a one‐way ANOVA with Tukey’s post‐test was performed. The number of independent, biological replicates varied between 5 and 23. ****P < 0.0001; ***P = 0.0004; ns, not significant. e Model for THPP activation of parkin with a defective Ubl domain.
Without CCCP, there was no ubiquitination of the Mfn2 substrate due to the absence of PINK1 and pUb (Fig. 6b). In contrast, CCCP-treatment led to an accumulation of PINK1, which led to parkin recruitment, phosphorylation/activation, and Mfn2 ubiquitination. In both conditions, addition of BIO-2007817 had no effect, in agreement with autoubiquitination assays with phospho-parkin (Fig. 1) and previous mitochondrial translocation and mitophagy assays48. BIO-2007817 was able to partially rescue Mfn2 ubiquitination by non-phosphorylatable parkin variants, either S65A or R0RBR, which lacks the Ubl domain (Fig. 6b). At 10 µM BIO-2007817, the fraction of total ubiquitination for Mfn2 was roughly 50% that of wild-type parkin.
We next asked if BIO-2007817 could rescue pathological mutations that impair parkin activity. R42P and V56E both affect the Ubl domain to destabilize its folding53,54 and impair mitophagy in cells in response to CCCP treatment35. BIO-2007817 at 10 µM effectively suppressed the ubiquitination defect of both mutations (Fig. 6c). The suppression was limited to Ubl mutations: neither R275W nor K211N showed enhanced activity in the presence of BIO-2007817. R275W occurs in the RING1 domain, disrupting parkin stability and possibly pUb binding35,55. The lack of effect on the K211N mutant recapitulates the conclusions of autoubiquitination and Ub-VS assays that pUb-binding is essential for activation by BIO-2007817 (Figure 1). The rescue of Mfn2 ubiquitination was selective. We did not observe rescue of either R42P and V56E by the diastereomer BIO-200781848 that differs at only one chiral center (Fig. 1e). Taken together, the in organello results show that mutations (ΔUbl, R42P, V56E, S65A) that affect the parkin Ubl domain can be rescued by BIO-2007817.
We turned to mitoKeima assays to confirm this result and assess the ability of BIO-2007817 to derepress non-phosphorylatable parkin mutants which do not normally induce mitophagy in cells. The assay measures mitophagy using mitochondrially targeted fluorescent protein (mitoKeima) that shifts its excitation spectrum when mitochondria enter the acidic environment of lysosomes. Depolarization of mitochondria by CCCP led to robust mitophagy in cells expressing wild-type parkin with or without 1 h pre-exposure to BIO-2007817 (Fig. 6d). Cells expressing non-phosphorylatable S65A parkin or ∆Ubl parkin showed only partial mitophagy that was enhanced by BIO-2007817. PD mutations destabilizing parkin Ubl (R42P and V56E) and reducing mitophagy response to CCCP, could be rescued by pre-treatment with BIO-2007817. In agreement with our previous findings, the importance of RING0 pUb/pUbl binding site in BIO-2007817 activation of parkin was confirmed by the complete loss of mitophagy response in K211N mutant which could not be rescued by the compound.
Based on our results, we propose a mechanism of activation of parkin by BIO-2007817 or THPP compounds (Fig. 6e). In the case of parkin with impaired Ubl, the THPP compound can allosterically promote the binding of pUb onto RING0 through bridging between parkin and pUb. When parkin Ubl can be phosphorylated by PINK1, the THPP binding site is already occupied by the ACT following pUbl relocalization to RING0. This explains the lack of activation effect of THPP on wild-type parkin observed in in organello and mitophagy assays, as well as in previously reported cell assays48.