This research included cross-sectional studies on the human microbiome and experimental studies in animals.
Ethical declaration
The study involved the collection of urine and stool samples using anaerobic methods and was approved by the Institutional Review Board, Faculty of Medicine, Chulalongkorn University, Thailand (COA No. 0995/2022). The experiments were performed in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants. The animal study protocol was approved by the Chulalongkorn University Animal Care and Use Committee of the Faculty of Medicine, Chulalongkorn University, Thailand (CU-ACUC No. 004/2565). All experiments were performed according to the IACUC and ARRIVE guidelines and regulations.
Human participants
In this study, eight patients with calcium oxalate urolithiasis received surgical treatment at the Chulalongkorn Hospital Urological Surgery Unit and ten healthy participants were included. Urine samples from the morning, first voiding spot urine samples were obtained to assess creatinine, calcium, and magnesium levels using electrochemiluminescence (COBAS C6000, Roche, USA) and to measure oxalate and citrate by capillary electrophoresis (P/ACETM MDQ, Beckman Coulter, USA). Fecal samples were also gathered using the AnaeroPouch -Anaero system (Mitsubishi Gas Chemical Co., Japan), from which a portion was treated to isolate DNA with a DNA/RNA shield (Zymo Research, USA) for subsequent sequencing.
16 S rRNA amplicon sequencing and analysis
To sequence 16s rRNA, the hypervariable V4 regions of the 16 S rRNA genes were amplified from DNA samples using 515 F (5′GTGCCAGCMGCCGCGGTAA 3′) and 806R (5′GGACTACHVGGGTWTCTAAT3′) primers and 2X KAPA hot start ready mix. The PCR conditions included an initial denaturation at 94 °C for 3 min, followed by 25 cycles of 98 °C for 20 s, 55 °C for 30 s 72 °C for 30 s, and a final extension step at 72 °C for 5 min. The 16 S amplicons were purified using AMPure XP beads and indexed using the Nextera XT index kit, followed by 8 cycles of the aforementioned PCR condition. Finally, the PCR products were cleaned and pooled for cluster generation and 250 bp paired-end read sequencing on the Illumina® MiSeq (Mod Gut Co., Ltd., Bangkok, Thailand).
The quality of the raw data was assessed using FastQC (v0.11.8) and MultiQC (v1.7). QIIME2 (version 2022.2) was applied to perform microbiome analysis. The adapter and preceding bases were trimmed at the 5′ end of the reads using the q2-cutadapt plugin. Reads with expected errors (maxEE) higher than 3.0 were discarded, paired end reads were merged with an overlap of 25 nucleotides between forward and reverse reads, and chimeric sequences were filtered using the q2-dada2 plugin.
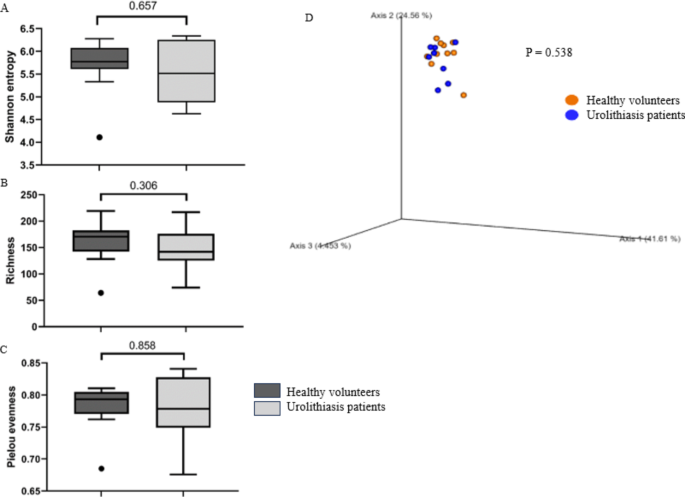
The taxonomic classifier was trained using the V4 sequence based on the naive Bayes classifier model using the q2-feature classifier plugin. Amplicon sequence variants (ASVs) were assigned for taxonomy based on the reference sequences of Sklearn method against the SILVA (version 138.1) 99% operational taxonomic units (OTUs). The unassigned mitochondria and chloroplast sequences were removed using the q2-taxa plugin. Calculations of the rarefaction curve, richness index diversity index, and Principal coordinate analysis (PCoA) and permutational multivariate analysis of variance (PERMANOVA) based on the Bray-Curtis distance were generated using the vegan package of R4.0.3.
Human urinary supersaturation index was calculated using the following equation16:
$$\:{\text{A}\text{P}}_{\text{C}\text{a}\text{O}\text{x}}=\:\frac{2.7\:\bullet\:{\:\text{C}\text{a}\text{l}\text{c}\text{i}\text{u}\text{m}}^{0.84}\:\bullet\:\:\text{O}\text{x}\text{a}\text{l}\text{a}\text{t}\text{e}}{{\text{C}\text{i}\text{t}\text{r}\text{a}\text{t}\text{e}}^{0.22}\:\bullet\:\:{\text{M}\text{a}\text{g}\text{n}\text{e}\text{s}\text{i}\text{u}\text{m}}^{0.12\:}\bullet\:\:{\text{V}\text{o}\text{l}\text{u}\text{m}\text{e}}^{1.03}}$$
APCaOx: Approximate Estimation of the Ion Activity Product of calcium oxalate.
Fecal microbiota transplantation (FMT) in Wistar rats
Three-week-old male Wistar rats, sourced from Nomura Siam Co. LTD in Thailand underwent a one-week acclimatization period before being housed in normal showbox cages in the Animal Lab at the Faculty of Medicine, Chulalongkorn University. The laboratory environment was meticulously controlled, maintaining a temperature of 25 °C, relative humidity between 30 and 50%, and a consistent 12-hour light/dark cycle. Throughout the study, rats had unrestricted access to food and water. To initiate partial eradication of the gut microbiota, their drinking water was supplemented with a cocktail of antibiotics (ampicillin 1 g/L, ciprofloxacin 1 g/L metronidazole 1 g/L, and vancomycin 0.5 g/L) for a duration of 7 days17. Following this phase, the rats were organized into two groups, each comprising six individuals. From week 0, systematic collections of blood, urine, and fecal samples were carried out for further analysis.
To create the fecal microbiota extract, we pooled 3 g of each fecal sample from urolithiasis into a urolithiasis-FMT mixture, and each sample from the volunteer into a healthy mixture of FMT, separately. Each sample was dissolved in 45 mL of 0.9% sodium chloride solution. The samples were then vigorously vortexed for 10 s and meticulously filtered three times through gauze, all under strict anaerobic conditions to preserve microbial integrity. Following filtration, the mixtures were centrifuged at 6,000 g for 15 min, allowing the collection of the clear supernatant18. The supernatant was then carefully combined with 20% glycerol, forming a protective medium for the microbial content, and subsequently stored at -20 °C for future analyses19.
During the experimental phase, the control group received an FMT solution derived from healthy individuals, while the urolithiasis group was administered FMT derived from patients with kidney stones. Each administration of FMT involved a 400 µL volume, administered by gavage twice weekly for a duration of four weeks. To simulate a high oxalate intake, sodium oxalate was incorporated into the drinking water of both groups at a concentration of 1% (w/w).
At the end of the experiment, the rats were placed in a metabolic cage for 24 h to collect the urine sample. Isoflurane was used as an anesthetic drug for blood collection from the retrobulbar vein before euthanasia using CO2 inhalation, and intestinal fecal samples were harvested immediately after euthanization. Subsequently, the intestines and jejunum were meticulously harvested for further analysis. In our biochemical assessment, blood samples were analysed for calcium and magnesium levels, while urine samples were evaluated for concentrations of calcium, magnesium, oxalate, citrate, creatinine and urinary supersaturation index.
Urinary supersaturation index was calculated using the following equation16:
$$\:{\text{A}\text{P}}_{\text{C}\text{a}\text{O}\text{x}}=\:\frac{4067\:\bullet\:\:{\text{C}\text{a}\text{l}\text{c}\text{i}\text{u}\text{m}}^{0.93}\:\bullet\:\:{\text{O}\text{x}\text{a}\text{l}\text{a}\text{t}\text{e}}^{0.96}}{{(\text{C}\text{i}\text{t}\text{r}\text{a}\text{t}\text{e}+0.015)}^{0.60}\bullet\:{\text{M}\text{a}\text{g}\text{n}\text{e}\text{s}\text{i}\text{u}\text{m}}^{0.55}\bullet\:{\text{V}\text{o}\text{l}\text{u}\text{m}\text{e}}^{0.99}}$$
For histological and genetic analyses, the intestines and kidneys were sectioned into two parts. The first section was forwarded to the Department of Pathology of the Faculty of Medicine, Chulalongkorn University, for detailed histopathological examination. The second section was RNA extraction using TRIzol LS reagent to facilitate RT-PCR assays. These assays were specifically designed to quantify the expression levels of intestinal zonula occluden-1 (ZO-1) and the oxalate transporter, Solute Carrier Family 26 Member 6 (SLC26A6), which offers insight into the molecular impact of our experimental interventions.
Fecal microbiota analyses
DNA extractions from bacterial feces were performed in both human and animal models using the Quick-DNATM Fecal/Soil Microbe Microprep Kit. The DNA extract was forwarded to ModGut Co., Ltd. (Thailand) for a comprehensive analysis of the fecal microbiota present in participants and animal models alike. To analyze the biological data of the 16 S rRNA (V3-V4 region), we employed QIIME software (Quantitative Insights into Microbial Ecology) software, a powerful tool for microbial community analysis. Sequences had been classified into various operational taxonomic units (OTUs), providing a detailed framework for understanding the diversity of microbials. To evaluate the diversity within the microbial population, we calculated the relative abundance using Shannon’s index.
Histopathology analyses
Kidney and jejunum samples were meticulously sectioned transversely, then carefully placed on cassettes and submerged in 10% neutral buffer formalin to ensure their preservation. The Department of Pathology of the Faculty of Medicine, Chulalongkorn University carried out the tissue staining process. The jejunum sections were stained with hematoxylin and eosin (H&E) and further analyzed through immunohistochemistry to detect ZO-1 expression.
For the assessment of histopathological changes in the kidneys of animal models, crystal deposits were examined in three distinct regions: the cortex, the cortical medullary junction, and the tip of the papillary21. This examination was conducted using a polarizing microscope at a magnification of X400. In a similar vein, jejunum sections were scrutinized under a microscope to evaluate ZO-1 staining, providing insight into the integrity of the intestinal barrier22. A pathologist was responsible for the scoring of these observations, ensuring a thorough and expert analysis of the histopathological features presented in both the kidney and intestinal samples of animal models.
Gene expression
Gene expression levels were accurately quantified using the quantitative polymerase chain reaction (qPCR) technique, employing the state-of-the-art QuantStudioTM 5 Real-Time PCR System. Complementary DNA (cDNA), synthesized from jejunum samples using the RevertAid First Strand cDNA Synthesis Kit, served as the template for measuring the expression of key genes: Zonula occluden-1 (ZO-1)23, Solute carrier family 26, Member 6 (SLC26A6)24 in the jejunum and the Nuclear Factor Kappa B subunit 1 gene (NF-kB)25 in the kidney. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as the reference gene23. CT values obtained from qPCR were utilized to calculate the gene expression levels using the 2−ΔΔCq method. This analysis could provide information on the relative expression levels of target genes compared to the housekeeping gene.
Statistical analysis
Data from research participants were analyzed using unpaired t-tests to compare general information, such as age. The chi-square test was used to analyze non-numeric data, such as gender. For the data from experimental animals, multiple repeated measures ANOVA with Bonferroni post hoc analysis was used to compare changes before and after the various values of the parameters. Additionally, Student’s t-tests were conducted to compare groups of rats, and paired t-tests were employed to analyze data with normal distributed, paired measurements, while Wilcoxon matched pairs signed rank test was used to evaluate unequal distributed data such as urinary mineral excretion. In the gut microbiome analysis, the most abundant taxa at the phylum and genus levels were demonstrated at the ASV level. The Mann Whitney test was used to evaluate the relative bacterial species. The Kruskal-Wallis test was employed for alpha and beta diversity analysis. Statistical analyses were performed using SPSS version 23.0 (IBM Statistics, USA) and GraphPad Prism version 9.4 (DotMatics, USA). The statistically significant was obtained when p < 0.05. The Chi-square test was utilised to analyze the tissue results.