Bacteria growth conditions
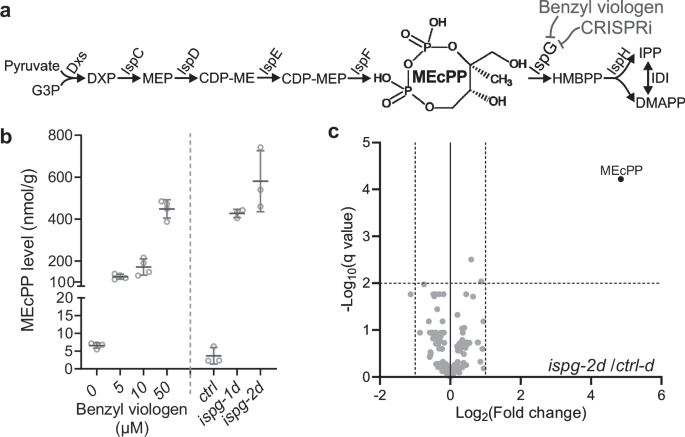
E. coli MG1655 Z1 malE– (F–, lambda–, rph-1, lacIq, PN25tetR, SpR, malE–)47 is acquired from Addgene (a gift from Keith Tyo, Addgene plasmid # 65915), herein referred to as the MG1655 strain. Unless stated otherwise, all bacteria were cultured at 37 °C in Luria Broth (Miller) (Sigma, L3522, USA) liquid medium or LB plates supplemented with 1.5% agar. The concentration of antibiotics for antibiotic-resistance selection were as follows: carbenicillin, 100 mg/L; kanamycin, 50 mg/L; gentamycin, 20 mg/L; chloramphenicol, 35 mg/L. A stock solution of 100 mM benzyl viologen (BV) was always freshly prepared and diluted into an LB medium to the desired concentration.
CRISPR interference-mediated knock-down of ispG in E. coli
Two sgRNAs targeting the non-template strand of 5’-end ispG coding region are designed using the Chop-chop website48. A control sgRNA (ctrl-d) is also generated by introducing two mismatch nucleotides in the ispg-2d sgRNA spacer region immediately adjacent to the protospacer adjacent motif (PAM), as diminishing the binding of dCAS9 to the target DNA strand. Incorporation of the above sgRNAs into the pgRNA vector of the CRISPRi system is performed as described49,50. Finally, the pgRNA and pdCAS9 vectors were transformed into E. coli MG1655 Z1 malE– strain by electroporation.
Plasmid constructs and primers
Sources of used plasmids are listed in Supplementary Table 1. All plasmids generated in this study, along with the primers and assembly methods, are listed in Supplementary Table 1. The E. coli strain for plasmid propagations is TOP10 (Invitrogen, ThermoFisher, USA). All primers used in the current studies are listed and annotated in Supplementary Table 2.
LC-MS analysis of MEcPP levels
An 8 mL LB medium supplemented with appropriate antibiotics or indicated concentration of BV was inoculated with 32 µL overnight culture of CRISPRi strains or wild-type E. coli cells and was shaken in a 37 °C incubator shaker until OD600nm reached ~ 1.0. Bacteria pellet from 5 mL of the culture was collected by centrifuge at 2500 × g for 10 min at 4 °C. The pellet was then washed with ice-cold water and pelleted into a preweighed 1.5 mL microtube for measuring the fresh weight of the collected bacteria. MEcPP was then extracted from these weighed pellets according to the published method51. Quantification of MEcPP levels was conducted using our established method52 with 3–4 biological repeats for each genotype or BV treatment.
Targeted Metabolomics of the Central Carbon and Energy Metabolism (CCEM)
Five biological repeats of ispg-2d, ctrl-d strains were cultured in LB medium supplemented with appropriate antibiotics in a 37 °C incubator shaker till late-logarithmic phase (OD600nm ≈ 0.8). Bacteria pellets were collected and washed with deionized water by centrifugation at 4 °C, followed by lyophilization. Subsequently, 30 mg of lyophilized E. coli was extracted in a two-phase extraction as described53 using 900 µL of dichloromethane/ ethanol (2:1, v/v) and 150 µL of aqueous hydrochloric acid of pH 1.4. After two rounds of extraction, aqueous extracts were combined and stored at − 80 °C until analysis. Separation of hydrophilic metabolites was performed by ion-pairing chromatography on a Nucleoshell RP18 column (2.1 × 150 mm, particle size 2.1 µm, Macherey & Nagel, GmbH, Düren, Germany) using a Waters ACQUITY UPLC System, equipped with an ACQUITY Binary Solvent Manager and ACQUITY Sample Manager (5 µL injection volume; Waters GmbH, Eschborn, Germany). Eluents A and B were aqueous 10 mM tributyl amine (adjusted to pH 6.2 with glacial acetic acid) and acetonitrile, respectively. Elution was performed isocratically for 2 min with 2% eluent B, from 2 to 18 min with a linear gradient up to 36% B, and from 18–21 min up to 95% B, and isocratically from 21 min to 22.5 min with 95% B, from 22.51 to 26 min again down to 2% B. The flow rate was set to 400 µL/min, and the column temperature was maintained at 40 °C.
Mass spectrometric analyses of small molecules were performed by targeted MS/MS via multiple reaction monitoring (MRM) by using a QTRAP 6500 (AB Sciex GmbH, Darmstadt, Germany) operating in negative ionization mode and controlled by Analyst 1.7.1 (AB Sciex GmbH, Darmstadt, Germany) (Supplementary Data 2). The source operation parameters were the following: ion spray voltage, − 4500 V; nebulizing gas, 60 psi; source temperature, 450 °C; drying gas, 70 psi; curtain gas, 35 psi.
Peak integration was performed using the MultiQuant software version 3.0.3 (Sciex, Toronto, CA). Individual CCEM peak areas of n = 5 biological replicates were normalized to the total peak area for each sample and averaged for the ispg-2d and the ctrl-d group, respectively. Finally, the group average of ispg-2d was divided by the average ctrl-d control samples. All area ratio data were logarithmized to the basis of 2.
RNA-Seq
Total RNAs from 37 °C shaking cultured E. coli cells of ispg-2d and ctrl-d strains (at late-logarithmic phase, OD600nm ≈ 1.0, three biological replicates for each genotype) were extracted using an AurumTM total RNA mini kit (Bio-Rad, USA). Genomic DNAs were removed from the total RNAs using a TURBO DNA-free Kit (Thermo Fisher, USA). Subsequently, ribosomal RNAs were removed using NEBNext rRNA depletion Kit (Bacteria) (NEB, USA), followed by RNA-Seq library preparation using NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB, USA). The quality of all RNA samples and prepared RNA-seq libraries were assessed using Agilent 2100 Bioanalyzer. The RNA-seq was conducted with an Illumina NovaSeq 6000 system on a S4 flow cell. HISAT254 was used to align sequencing reads to the E. coli reference genome. DESeq255 was used to count and normalize mapped reads. Genes with 2-fold altered expression levels and q-value ≤ 0.05 (unpaired t tests) were identified as differentially expressed genes.
Biofilm culture and visualization
Biofilm production was quantified using crystal violet staining, and statistical significance was determined using Brown-Forsythe and Welch ANOVA and Dunnett’s multiple comparisons tests (CRISPRi strains) and RM one-way ANOVA tests with the Geisser-Greenhouse correction and Dunnett’s multiple comparisons tests (BV treatment), with p < 0.05 considered statistically significant. Specifically, overnight cultured E. coli cells were 1:100 diluted into fresh LB medium with appropriate antibiotics in 96-well non-treated polystyrene (PS) plates (Greiner, Germany) or polyvinyl chloride (PVC) “U” bottom plates (Corning, USA). After 48 h of static culture at room temperature (22–23 °C), OD600nm of the bacterial cultures were measured on a Molecular Devices SpectraMax iD5 plate reader, followed by crystal violet staining of biofilm as previously described56. Stained biofilm was dissolved in 30% acetic acid and quantified by measuring OD570 nm on the plate reader.
AFM imaging of E. coli fimbriae
Overnight-grown E. coli cells were 1:2000 diluted into fresh LB medium with appropriate antibiotics or indicated concentrations of BV and statically cultured in a 37 °C incubator for 24 h. The culture at medium-air interface was transferred onto poly-L-lysine-coated cover glasses and incubated at room temperature for 1 h for adhesion of bacteria cells to coverglass. Subsequently, the cover glasses were washed three times by dipping into a tank of deionized water. The cover glasses were then air-dried and mounted on a JPK NanoWizard 4a atomic force microscope (Bruker, USA) for imaging with AC mode using a Bruker RFESPG-75 or µMasch HQ:NSC18/Al BS cantilevers.
Sample preparation for LiP-MS proteome analyses
Three biological repeats of ispg-2d, ctrl-d, and MG1655 strain were cultured in LB medium supplemented with appropriate antibiotics in a 37 °C incubator shaker till late-logarithmic phase (OD600nm ≈ 0.8). Total protein extraction was prepared according to the previously described57. Total protein extraction from ispg-2d and ctrl-d was directly subject to LiP-MS assay57. For exogenous MEcPP or MEP treatment, total protein extraction from MG1655 was incubated with 200 µM MEcPP or MEP at room temperature for 30 min, then was used for LiP-MS assays57. An equal input of 100 μg of all extracted or MEcPP/MEP-treated protein samples was subject to limited proteolysis (LiP) using proteinase K followed by trypsin/Lys-C digestion or directly digested by trypsin/Lys-C. The abundance of unique peptide features was quantified in the LiP samples, while protein abundance was quantified in the sample digested with only trypsin/Lys-C, which was then used for normalization of the unique peptide abundance in the LiP samples. The protein samples were processed for proteomic analysis following the established method57. Briefly, the samples from LiP-MS assays were dissolved in a denaturation buffer (6 M urea, 2 M thiourea dissolved in 50 mM ammonium bicarbonate) followed by reduction with 100 μM DTT for 60 min at room temperature. The alkylation step was done by 60 minutes of incubation with 300 μM iodoacetamide in the dark. Residual iodoacetamide was quenched by 100 μM DTT for 10 min at room temperature. The residual urea is diluted with at least 6 volumes of 50 mM ammonium bicarbonate. Finally, LiP-MS fractions were digested using trypsin/Lys-C mixture (Mass Spec Grade, Promega) for 16 h according to the manufacturer’s instruction. Digested peptide samples were desalted using C18 sep-pak column plates. The eluted peptides were transferred to Eppendorf low-bind tubes and then dried using speed vac. The dried peptides were resuspended using a resuspension buffer (5% acetonitrile in 0.1% formic acid). An equal amount of the peptides was injected for LC-MS/MS analysis.
Data acquisition for LiP-MS proteome analyses
The peptide mixtures were separated using a Dionex-ultimate 3000 nano-liquid-chromatography system (nHPLC) with an acclaim pepmap C18 column. A flow rate of 300 nL/min was used for the separation of complex peptides. The solvent A/B gradient was as follows: Being isocratic at 3% B for 17 min (including the first 10 min for loading and trap column), linearly increasing to 30% B at 125 min, linearly increasing to 40% B at 145 min, keeping at 95% B from 145.1 min to 155 min, shifting back to 3% B in 0.1 min (valve were returned to load position) and holding until 170 min. The peptide samples were sprayed using a nano-bore stainless-steel emitter (Fisher Scientific). Peptides were analyzed using an Orbitrap-Exploris-480 MS™ mass spectrometer. Data was collected using a data-dependent acquisition (DDA) mode. Standard mass spectrometer parameters were kept as described58, HeLa digests (Pierce, Thermo Scientific, USA) have been used to monitor the retention time drift and mass accuracy of the instrument.
LiP-MS proteome data process
Raw data were analyzed using the Proteome Discoverer (PD) (version 3.0, ThermoFisher Scientific) following the manufacturer’s instructions. PD search was made using an E. coli protein database (UP000000625, Escherichia coli (strain K12) (K12/MG1655/ATCC 47076)) downloaded from Uniport. The trypsin enzyme was selected and used in semi settings. Common contaminants were compiled and added to the search. SEQUEST HT was used to assign the peptides, allowing a maximum of 2 missed tryptic cleavages. In addition, a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.02 Da, with a minimum peptide length of 6 AAs, were selected. Cysteine-carbamidomethylation and methionine-oxidation were selected in default modifications. Label-free quantification (LFQ) based on MS1 precursor ion intensity was performed in Proteome Discoverer with a minimum Quan value threshold set to 0.0001 for unique peptides. The ‘3 Top N’ peptides were used for area calculation. Quality control plots for the LiP-MS analyses are provided in Supplementary Data 7.
Recombinant protein expression
Cloning (pET16b-IspC), expression, and purification of 6xHis-tagged IspC protein were previously described59. The coding sequence of ispG and hns were cloned into the pET-28a vector for expression of 6xHis-tagged recombinant protein in E. coli Rosetta (DE3) cells. Recombinant protein production is induced with 0.1 mM Isopropyl β-d-1-thiogalactopyranoside (IPTG) at room temperature for 4 h and affinity purified with TALON Metal Affinity Resin (Takara Bio, USA). Eluted recombinant proteins (elution buffer: 50 mM sodium phosphate buffer pH7.4, 300 mM NaCl, 100 mM imidazole) were buffer exchanged using Amicon Ultra-15 centrifugal filters (MilliporeSigma, USA) to remove imidazole. Eventually, 6xHis-H-NS is stored in 50 mM Tris-Cl pH7.5, 200 mM NaCl, 10% (v/v) glycerol, and 6xHis-IspG in 50 mM Tris-Cl pH7.5, 50 mM NaCl, 10% (v/v) glycerol. All recombinant proteins are stored as 30–50 µL aliquots in 200 µL microtubes and kept in a −80 °C freezer after snap-freezing in liquid nitrogen.
Protein thermal shift assay
Protein thermal shift assay was performed in a Bio-Rad CFX96 Real-Time PCR detection system using a Protein Thermal Shift Dye Kit (ThermoFisher, USA). Thermal shift buffer for each recombinant protein was as follows: IspC, 25 mM PIPES pH7.0, 500 mM NaCl; IspG, 25 mM MOPS pH7.5, 50 mM NaCl; H-NS, 25 mM PIPES pH6.0, 50 mM NaCl. Each reaction (20 µL) contains 1x thermal shift dye, 2 µM recombinant protein, and 200 µM of MEcPP, HMBPP or NH4OAc.
Microscale thermophoresis
His-tagged H-NS protein was labeled with the RED-MALEIMIDE dye according to the manufacturer’s instructions (Nanotemper, Germany). Binding was performed in standard capillaries in 25 mM MES pH 6.0 and 50 mM NaCl. MST power was set to high. Data are the mean of eight independent measurements from four independent titrations. His-tagged IspG protein was labeled with the His-Tag Labeling Kit RED-tris-NTA 2nd Generation dye according to the manufacturer’s instruction (Nanotemper, Germany). Binding was performed in standard capillaries in 25 mM MOPS pH 7.5 and 50 mM NaCl. MST power was set to medium. Data are from four independent titrations. Data analysis was performed using Monolith software (Nanotemper, Germany).
Electrophoresis mobility shift assay (EMSA)
DNA probe of fimE promoter for the EMSA was PCR amplified using Q5 High-Fidelity DNA polymerase (NEB, USA) and purified with a HighPrep PCR kit (MagBio Genomics, USA). In a 20 µL reaction, 240 nM 6xHis-H-NS was first mixed with increasing concentrations of MEcPP or MEP in a Tris buffer (10 mM Tris-Cl pH7.5, 10 mM MgCl2, 10 mM KCl, 100 mM NaCl, 0.5 mM DTT, 5% glycerol, 0.1% (w/v) BSA, and 1 mM spermidine) and kept at 25 °C for 20 min on a Bio-Rad C1000 Touch Thermal Cycler. The reaction was incubated for another 20 min at 25 °C after the addition of 30 nM fimE promoter DNA fragments. The reaction mix was resolved by running in a 5% polyacrylamide gel (1xTAE) at 100 V. The gel was imaged in a Bio-Rad ChemiDoc MP Imaging System after staining in GelRed (Biotium, USA) staining solution (1xGelRed in 1xTAE, 100 mM NaCl) for 30 min.
Screening of biofilm revertant after random transposon-insertion mutagenesis
Transposon-insertion mutant library of ispg-2 strain was generated by mutagenesis plasmid pSNC-mTn519. More than 9150 individual mutant clones were cultured and tested for biofilm formation. All biofilm revertants from the first-round screening were further validated for at least 3 more times which resulted in 265 mutant clones consistently recovered biofilm formation.
Mapping transposon insertion site
An arbitrarily primed PCR approach60 followed by Sanger sequencing was used to acquire the flanking sequences of the Tn5 transposon insertion site in the biofilm revertants. An initial examination of 7 randomly selected revertant clones has led to the identification of an ispg-2 fimE::Tn5 mutant.
Homolog recombination mediated fimE knockout
Kanamycin resistant cassette was PCR-amplified from plasmid pDONR221 using Q5 High-Fidelity DNA polymerase (NEB, USA) with primers designed with 5’ extensions homologous to the sequences flanking the coding region of fimE. Homolog recombination mediated fimE knockout in ispg-2 strain using plasmid pSIJ8-Gent61 was performed as described61,62.
Reverse transcription and quantitative PCR (RT-qPCR)
Overnight cultures of CRISPRi strains (ispg-2 and ctrl), the revertant ispg-2 fimE::Tn5, and MG1655 strain were 1:2000 diluted into 3 mL LB liquid medium with appropriate antibiotics or indicated concentrations of BV and were statically cultured at 37 °C for 24 h. Total RNAs from the aforementioned E. coli cells were extracted using a Bio-Rad Aurum total RNA mini kit. Genomic DNAs were removed from the extracted RNAs using a TURBO DNA-free Kit (Invitrogen, Thermo Scientific, USA). Complete removal of genomic DNA contamination from the treated RNA samples was verified by the absence of PCR amplification using qPCR primers targeting era genes in the non-RT reactions. Subsequently, the RNAs were reverse transcribed using iScript™ Reverse Transcription Supermix (Bio-Rad, USA). To find the most stable reference genes for QPCR, we performed QPCR reactions using qPCR primer pairs for 7 reference genes (secA, hcaT, era, dnaG, cysG, gyrB, and idnT) on the RT products of 6 representative samples (include various genotypes and BV-treatments). The qPCR results were subjected to geNorm analysis in qBase + (v3.4) software63, which recommended an optimal normalization factor to be calculated using the geometric means of two reference genes, hcaT and cysG. Finally, qPCR of ispG, fimE, hcaT, and cysG was performed on all RT samples, and the resulting Cq value was analyzed by the qBase+ software, which generated normalized RNA level of ispG and fimE in the samples. Each genotype or BV treatment has at least three biological replicates and two qPCR technical replicates.
Analysis of fimA promoter inversion
Analysis of the fimA promoter inversion was performed as previously described37. The promoter of fimA was amplified using NEB Taq DNA polymerase and purified using a HighPrep PCR kit (MagBio Genomics, USA). Purified fimA promoter fragments were digested with HinfI overnight at 37 °C and then resolved by running on a 5% polyacrylamide 1xTAE gel at 100 V. The gel was stained with 1x GelRed staining solution and imaged on a Bio-Rad ChemiDoc MP Imaging System.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.