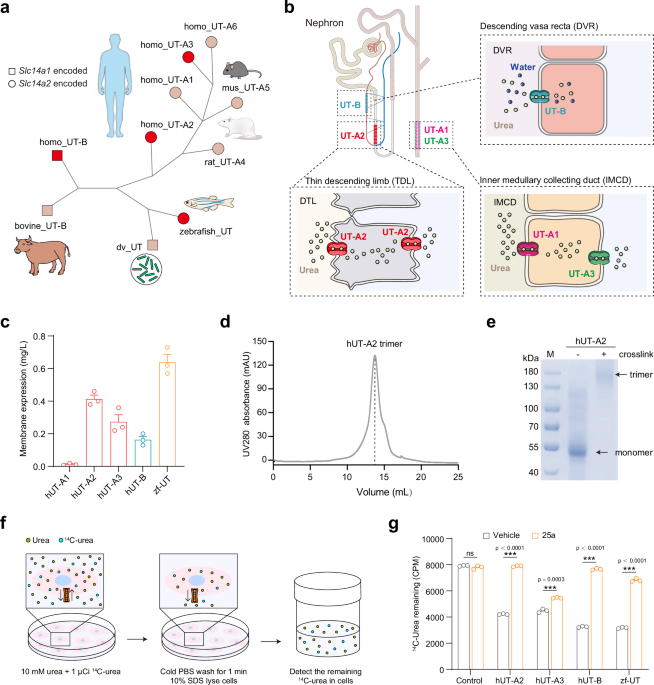
Expression and functional characterization of human UT
To facilitate the structural determination of UTs, we screened the plasma membrane expression levels of UT proteins. Flag tags were added to the N-termini of zebrafish UT (zf-UT) and human UT-A1, A2, A3, and UT-B sequences, and the mCherry protein was fused by the TEV cleavage site and GSA linker at the C-termini. zf-UT had the highest plasma membrane expression, followed by hUT-A2, hUT-A3 and hUT-B, whereas hUT-A1 showed no detectable expression in the insect cell membrane (Fig. 1c). Further examination of UT proteins by size exclusion column chromatography showed that the elution volumes of four highly expressed UT proteins (approximately 14.5 ml in Sepharose 6) were significantly earlier than those of the expected monomer proteins (approximately 17 ml), suggesting that these four UT proteins existed in the multimer forms (Fig. 1d and Supplementary Fig. 1b–d). Cross-linking with glutaraldehyde followed by gel electrophoresis suggested that the four highly expressed UT proteins all existed as trimers (Fig. 1e). These observations recalled previous reports of the trimer formation of Desulfovibrio vulgaris UT (dv_UT) and bovine UT-B1,19. Importantly, urea permeability, measured using the urease assay and 14C-labeled urea transport assay, was significantly higher in HEK293 cells transfected with zf-UT, hUT-A2, hUT-A3, and hUT-B than in HEK293 cells transfected with the control pcDNA3.1 plasmid (Fig. 1f, g and Supplementary Fig. 1e, f). Moreover, the urea permeability of hUT-A2, hUT-A3, hUT-B, and zf-UT was blocked by the small-molecule inhibitor 25a (Fig. 1f, g).
Overall cryo-EM structures of the UT trimers
We then determined nine structures of hUT-A2 and zf-UT in the resting state and bound to endogenous urea, the competitive inhibitor 25a or ATB3, the uncompetitive inhibitor CF11, or the noncompetitive inhibitor HQA2 via single-particle cryo-electron microscopy at resolutions ranging from 2.6 to 3.3 Å (Fig. 2a, d and Supplementary Figs. 2–5 and Supplementary Tables 1 and 2). The cryo-EM structures of hUT-A3 and hUT-B in the absence of urea were also solved at resolutions of 2.3 Å and 2.4 Å (Fig. 2b, c and Supplementary Fig. 4 and Supplementary Table 2). The cryo-EM densities allowed the assignment of most residues of the UT proteins, including residues 51 to 381 of hUT-A2, residues 96 to 440 of hUT-A3, residues 34 to 383 of hUT-B and residues 31 to 380 of zf-UT (Supplementary Table 3). The density of urea and inhibitors could also be assigned in the hUT-A2 and zf-UT structures in either intracellular or extracellular regions of the transport channels (Fig. 2a, d).
a–d Overall cryo-EM densities and structures of hUT-A2 (a), hUT-A3 (b), hUT-B (c) and zf-UT (d) homotrimer. The cryo-EM densities of different UTs are displayed on the phospholipid bilayer cell membrane with the represent cartoon models shown below. Molecules bound to the extracellular and intracellular sides of different UTs, along with their corresponding EM densities, are indicated in dashed circles at the top and bottom layers, respectively. Created in BioRender. Huang, S. (2024) https://BioRender.com/h99o001.
Consistent with the electrophoresis results, zf-UT, hUT-A2, hUT-A3 and hUT-B all existed as homologous trimers in the solved cryo-EM structures (Fig. 2 and Supplementary Fig. 6a–d). In all these UT structures, each UT unit consists of 10 complete transmembrane helices and 2 semi-transmembrane helices distributed symmetrically along the axis of the urea transport channel, which were named 1a–5a, 1b–5b, and Pa, and Pb, respectively (Supplementary Fig. 6e). The two symmetric halves of each UT subunit are named the a-half and b-half, respectively, and are connected by a long extracellular loop (LECL) (Supplementary Fig. 6e). The urea transport channels of all UTs are enclosed by helices from both halves, including helices 3a, 3b, 5a, 5b, Pa, and Pb (Fig. 3a and Supplementary Fig. 6a–e)1,19. Both the N-terminus and C-terminus of each UT subunit face the intracellular side. A portion of the C-terminus of UT folds into a helical-like structure and is named the C1 helix. Between the C1 helix and helix 5b is the long intracellular loop (LICL), which is also located on the cytoplasmic side (Supplementary Fig. 6e).

a The urea transport channel of hUT-A2 is shown as blue dots with the surrounding helices displayed as cylindrites. The urea transport channel of hUT-A2 is a continuous cavity with a length of approximately 21 Å which enclosed by helices from both halves, including helices 3a, 3b, 5a, 5b, Pa, and Pb. b The EM density in the urea binding pocket of hUT-A2 at the 4.0 σ contour level are shown as red mesh on both the extracellular and cytoplasmic sides of the urea transport channel. c The extracellular urea binding pocket (EUBP) and the cytoplasmic urea binding pocket (CUBP) of hUT-A2. d, e. The key residues interacting with urea in EUBP (d) and CUBP (e) are shown as sticks while the urea and water molecules are displayed as stick-balls and red balls, respectively. The H-bond are shown as red dash line. f Structurally equivalent residues in the EUBP (left) and CUBP (right) of hUT-A2 compared with other hUTs and zf-UT. The secondary structure of the indicated residues is shown at the top. The conserved QPb-T5b-T5a-QPa motif of urea transport channel in different UTs is shown in blue backgrounds. g, h Urea permeability shown by the detection of remaining 14C-labeled urea in cell transfected with hUT-A2 (g) and zf-UT (h). Cells transfected with wild type and mutant UT plasmids are shown as gray and green columns respectively, while cells transfected with pcDNA3.1 blank plasmids for control are shown as blank column. Data are represented as mean ± SEM from 3 independent experiments (n = 3). ***P < 0.001, statistical differences were determined by the two-sided unpaired Student’s t-test (compared with wild type).
Importantly, most structural differences between different UTs are located in the N-terminus, C-terminus, LECL and long intracellular loop (LICL). Compared with the structure of hUT-A2, the N-terminus of hUT-A3 has an extra α helix, named the N1 helix. The C1 helix of hUT-A3 is one helical turn shorter than the C1 helix of hUT-A2 and is rotated outward by approximately 8° (Supplementary Fig. 6f). Compared with hUT-A2, the N-terminus of hUT-B has two more helical structures, which are named N1 and N2. The C1 helix of UT-B is rotated outward by approximately 11°, and the intracellular ends of helix 4a, LECL and LICL are shifted inward compared with the corresponding positions in UT-A2 (Supplementary Fig. 6g). The N-terminus of zf-UT is similar to the N-terminus of hUT-B, with two helices, N1 and the N2. Compared to hUT-A2, the C1 helix of zf-UT is rotated outward by approximately 6°, and the LECL and LICL are shifted inward (Supplementary Fig. 6h).
Capture of urea in the urea transport channel
In the hUT-A2 structure, the urea transport channel connecting the extracellular section to the intracellular region is composed of helices 3a, 3b, 5a, 5b, Pa, and Pb. The channel is a continuous cavity that is wide at both ends and narrow in the middle, with a length of approximately 21 Å (Fig. 3a). Compared with the apo-hUT-A2 structure, the additional EM density in this potential urea transport channel at the 4.0 σ contour level enabled us to model the urea molecule and its coupled water on both the extracellular and cytoplasmic sides (Fig. 3b and Supplementary Fig. 7a, b). The modeled urea molecules were further supported by molecular dynamics simulation (Supplementary Fig. 7c). There are two urea binding pockets in the channel: one in the extracellular urea binding pocket (EUBP) and the other in the cytoplasmic urea binding pocket (CUBP), separated by approximately 10 Å (Fig. 3c–e). In total, 7 residues compose the EUBP, and the largely hydrophobic environment is decorated with many polar groups. In the EUBP, two nitrogen atoms of the upper urea form a salt bridge with the main chain carbonyl oxygens of Q231Pb and V232Pb, a hydrogen bond with the side chain of T3385b and a water-mediated hydrogen bond with the side chain of Q231Pb (Fig. 3d). In addition to polar interactions, urea molecules form a cation-π interaction with the benzene ring of F2873b and van der Waals interactions with L1313a, F1795a, and C3375b (Fig. 3d).
The CUBP consists of 5 hydrophobic residues and 2 polar residues (Fig. 3e). Notably, the lower urea molecule in the CUBP form symmetric polar interactions and hydrophobic packing compared with the upper urea molecule in the EUBP, including interactions with the main chain carbonyl groups of Q67Pa and V68Pa and the side chain of T1765a, as well as cation-π interactions with F70Pa and Y1233a (Fig. 3e). The motif Q67PaV68PaY1233aT1765a in the CUBP and the motif Q231PbV/I232PbF/Y2873bT3385b in the EUBP are conserved among UT-A2, UT-A3, UT-B and zf-UT and thus could be a “urea recognition motif (URM)” in UTs (Fig. 3f). In addition, the lower urea molecule in the CUBP forms contacts with F1755a and F3415b (Fig. 3e). Consistent with these observations, alanine mutations of the residues in the URM of either the CUBP or the EUBP, such as Y1233a and T1765a in the CUBP or Q231Pb F/Y2873b and T3385b in the EUBP significantly decreased urea transport by these UTs (Fig. 3g, h and Supplementary Fig. 7d–g).
Potential urea permeation mechanism of the human urea transporter hUT-A2
Compared with that of apo-hUT-A2, the pore size of the urea transport channel of urea-bound hUT-A2 is significantly smaller, and the mean radius is decreased by approximately 0.3 Å (Fig. 4a and Supplementary Fig. 7h). The shrinkage of the channel radius is mainly due to the inward movements of residues such as F70Pa, Y1233a, L1273a, L1313a, F1795a and F2873b (Supplementary Fig. 7i). We speculated that the hydrogen bonding and hydrophobic interactions of urea in the urea transport channel are the driving forces for channel cavity shrinkage and the resulting smaller radius (Fig. 4b). The UT transport channels are divided into three parts, So, Sm, and Si, from the extracellular to the cytoplasmic side. In the So region, the urea molecule forms a salt bridge with the main chain oxygen atoms of Q231Pb and V232Pb and a hydrogen bond with the hydroxyl group of T3385b (Fig. 4b). The hydrophobic side chains of L1313a and F2873b provide steric hindrance for the orientation of the urea molecule. These interactions induce the inward movements of several residues inside the transport channel, such as L1273a, L1313a, and F1795a, as well as F2873b on the other side (Supplementary Fig. 7i).

a The urea transport channel of hUT-A2 is highlighted by blue dotted line with the inner urea molecules displayed as stick-balls. b The hydrogen bonding and hydrophobic interactions of urea in the urea transport channel of hUT-A2. The black dashed lines are used to separate So, Sm, and Si regions of the channel. The H-bond are shown as red dashed line and the cation-π interaction are shown as purple dashed lines. c Free energy surface (FES) of urea within the hUT-A2 channel. The simulation reveals three distinct low-energy states, which are characterized by prolonged urea residence times across defined regions within the channel and corresponds to the So, Sm, and Si regions respectively. d Simplified one-dimensional energy landscape highlighting the So, Sm, and Si states of urea within the hUT-A2 channel. e Metadynamics analysis depicts urea dynamically transitioning between three states-So, Sm, and Si-within the hUT-A2 channel, with distribution probabilities of 37%, 19%, and 34%, respectively. f–h Metadynamics simulations elucidate the passage of urea through the Sm region. This process involves a rotation of approximately 40°in the side chains of T1765a and T3385b, facilitating the rearrangement of T1765a–T3385b side chains (f). Notably, hydrogen bond formations between urea and specific residues, including T3385b and T1765a, are depicted (g). The rotation of side chains of T338/T176 and structural rearrangement of L127/L291 form a hydrophobic cavity in the Sm region, guiding urea transportation across the channel (h). The H-bond are shown as red dashed line. The blue arrows indicate the direction of movement of the urea molecule. i Urea permeability shown by the detection of remaining 14C-labeled urea in cell transfected with different hUT-A2. plasmids are shown as gray and green columns respectively, while cells transfected with pcDNA3.1 blank plasmids for control are shown as blank column. Data are represented as mean ± SEM from 3 independent experiments (n = 3). **P < 0.01, ***P < 0.001, statistical differences were determined by the two-sided unpaired Student’s t-test (compared with wild type). j Schematic diagram of urea permeation mechanism of hUT-A2.
The urea molecule in the Si region shows a similar symmetric bonding pattern. The Sm region of the UT transport channel is a relatively narrow cavity consisting of three residue pairs, namely, T1765a–T3385b, L1273a–L2913b, and F1793a–F3413b (Fig. 4b). The pore radius of this central core of the Sm region is significantly smaller than that of the So and Si regions (Supplementary Fig. 7h). Because our current cryo-EM data of urea-hUT-A2 could not capture clear EM density inside the Sm region, we performed MD simulation to explore the process of urea molecule passing through the Sm region (Supplementary Fig. 7j). The simulation captured three low-energy states with longer residence times of urea across the channel, located in the three defined regions, So, Sm, and Si, with distribution probabilities of time of 37%, 19% and 34%, respectively (Fig. 4c–e). According to kinetic simulation and cluster analysis, the side chains of T1765a and T3385b rotated by approximately 40° followed by the rearrangement of the side chains of T1765a–T3385b and F1793a–F3413b to allow the urea molecule to pass through the Sm region. In the Sm region, the oxygen atom of urea formed hydrogen bonds with the hydroxyl group of the T1765a-T3385b, while the nitrogen atom formed hydrogen bonds with the main chain carbonyl group of V68Pa-V232Pb (Fig. 4f, g). Moreover, the polar urea molecules also interacted with the hydrophobic side chains of T3385b/T1765a, V232Pb/V68Pa and L1273a/L2913b through van der Waals forces in the Sm region (Fig. 4g, h). The urea molecules were sterically restricted by the side chains of amino acids L1273a–L2913b and F1793a–F3413b in the Sm region (Fig. 4h). Therefore, urea molecules shuttle through the transport channel in the form of a “monolith”, supporting a previous hypothesis1,19. We speculated that the urea molecule rotated via H-bond formation with the Q231Pb–T3385b–T1765a–Q67Pa motif during transportation (Supplementary Movie). Consistent with these simulation results and our hypothesis, the mutations Q67A, T176A, Q231A and T338A etc. of hUT-A2 significantly impaired urea permeation efficiency (Figs. 3g, 4i and Supplementary Fig. 7k, l).
We therefore hypothesized that the transport of urea across hUT-A2 occurs in three steps. In the first step, free urea molecules in solution are captured by the flexible side chain of Q231Pb in the extracellular region. The urea molecules are then brought into the So region of the transport channel by the rotation of the side chain of Q231Pb and coordinated by water molecules. At this stage, the two basic nitrogen atoms of the urea molecule form a salt bridge with the negatively charged poles of the Pa helix backbone (carbonyl oxygen atoms of the Q231Pb and V232Pb main chains). The hydrophobic nature of L1313a–F2873b facilitates a defined orientation of the urea molecule on the horizontal plane. In the second step, the flexibility of the side chains of L1313a and F2873b and hydrogen bond transfer mediated by the side chain of T3385b pull the urea molecules into the Sm region. In particular, rearrangement of the configurations of the residue pairs L1273a–L2913b and F1793a–F3413b may allow the entry of urea molecules. In the third step, the movement of the T1765a side chain may transfer the urea molecules from the Sm region to the Si region, forming an interaction pattern that is symmetric to the interaction pattern of urea in the So region (Fig. 4j). The urea may then move downward to form a hydrogen bond with the side chain of Q67Pa. Finally, the urea bound to the Si region is released into the cytoplasm, thus completing the transport of urea from the extracellular to the intracellular space. We speculated that the formation and transfer of hydrogen bonds between urea and the Q231Pb–T3385b–T1765a–Q67Pa motif play important roles throughout the whole process of urea transport (Fig. 4j). Because the distribution of amino acids in the So and Si regions of the channels is relatively symmetrical, we speculated that urea molecules can be transported from the intracellular end to the extracellular end in a similar manner (Fig. 3f).
Observation of urea transport by zf-UT
The trimer structure of urea-bound zf-UT was solved at an overall resolution of 3.0 Å. Compared with the apo-zf-UT structure, obvious and continuous EM densities enabled easy assignment of urea on both the intracellular and extracellular sides of the zf-UT channel (Figs. 2d, 5a, b and Supplementary Fig. 8a–c). According to the EM density distribution, the urea transport channel contains a vertical tubular cavity composed of 3a, 3b, 5a, 5b, Pa, and Pb helices and then extends into the intracellular section composed of the intracellular ends of helices 3a–5a and ICL2b (Fig. 5c). Due to the continuous EM density, MD simulations were performed to model urea at specific positions of the urea transport channel (Fig. 5d–f).

a, b Overall view of the urea density in zf-UT trimer. Two portions of obvious and continuous cryo-EM densities corresponding to urea in zf-UT are located at both the intracellular and extracellular sides of the channel (a), which facilitated the unambiguous assignment of urea molecules (b). c Comparison of the transportation channel of zf-UT (blue) and hUT-A2 (green). According to the EM density distribution, the urea transportation channel of zf-UT contained a vertical tubular cavity composed by 3a, 3b, 5a, 5b, Pa, and Pb helices, and then extends into the intracellular section composed by intracellular ends of helices 3a–5a and ICL2b. d Free energy surface (FES) of urea across the zf-UT channel, revealing six low-energy states-So’, So, Sm, Si, Si’, and Si”-associated with extended urea residence times across six distinct regions within the channel. e Simplified one-dimensional energy landscape illustrating the So’, So, Sm, Si, Si’, and Si” states of urea within the zf-UT channel. f The location of the urea molecules are modeled into six regions shown as So’, So, Sm, Si, Si’, and Si” in the zf-UT channel surrounding with presentation of key interacting residues. The urea transport channel of zf-UT is depicted by white dotted line with the inner urea molecules displayed as stick-balls. The conserved QPb–T5b–T5a–QPa motif is shown as green dots. g, h The interactions between urea and the key residues in the zf-UT urea transport channel (shown as blue dots) in the extracellular side (g) and the intracellular side (h) of the channel. The H-bonds are shown as red dashed line. i Barcode comparisons of the residues involving in the urea channel of zf-UT between different UTs. The residues predicted to involve with common interactions with urea molecules are displayed as black circles, including residues of the QPb–T5b–T5a–QPa motif filled with green, while the residues predicted to form distinct interactions are shown as black circles filled with gray. The letters inside the circles represent different amino acids present in the corresponding positions.
Compared with the cryo-EM structure of urea-hUT-A2, the structure of urea-zf-UT has additional EM density connected to the “So” region on the extracellular side, which is named the “So’” region, a cavity created by the side chains of F1353a, L2843b, and P335ECL2b (Fig. 5f and Supplementary Fig. 8b–e). Our MD simulation indicated that F1353a and L2843b form transient interactions with urea during urea transport (Fig. 5f–h). The conservation of F1353a and L2843b among UTs of different species suggested that the channel governed by these two residues is a common path for urea transport (Fig. 5i). On the intracellular side, the EM density of zf-UT-urea enabled the identification of two other urea binding sites, which were named Si’ and Si”, connecting to the Si region via an “L” configuration (Fig. 5b, f, h and Supplementary Fig. 8c). Whereas urea in the Si’ region contacts F3003b and L363ICL2b, urea in the Si” region interacts with Y1213a and S1604a (Fig. 5h and Supplementary Fig. 8c). We noticed that key residues in the urea transport channel, such as Y2873b, L1203a, and Y1213a of zf-UT, were replaced by F, F, and H, respectively (Fig. 5i). These amino acid differences may contribute to the stronger and longer EM density of urea observed in the zf-UT-urea compared with that in the hUT-A2-urea complex structure.
In the So and Si regions of the channel, urea showed similar interaction modes to those in the urea-hUT-A2- structure, including the polar network constituted by the Q231Pb–T3385b–T1765a–Q67Pa motif and hydrophobic interactions involving L1313a, F2873b, F70Pa, and Y1233a, which are distributed along the channel from the extracellular side to the intracellular side (Fig. 5i). These results indicate that a common mechanism and conserved motifs underlie urea transport in UTs from different species.
Competitive inhibition of UT-A2 by 25a
25a is a synthetic competitive inhibitor of UT with potency in the micromolar range (IC50 = 0.58 ± 0.01 μM toward hUT-A2) (Fig. 6a and Supplementary Fig. 9a, Supplementary Table 4). We solved the cryo-EM structure of hUT-A2 with 25a to understand the structural basis of its inhibition and to facilitate more potent and selective UT inhibitor design. Unambiguous EM density corresponding to 25a could be easily assigned on both the extracellular and intracellular sides (Figs. 2a, 6b and Supplementary Fig. 9b). The furan group of 25a, which mimics the urea molecule, inserts deeply into the channel center and occupies the pocket region of the EUBP and CUBP (Fig. 6c–f and Supplementary Fig. 9c–f, Supplementary Table 5). On the extracellular side, the methyl ketone group and amide group on the two sides of furan form hydrogen bonds with the side chains of T338 and Q231 (Fig. 6c, e). In addition to polar interactions, the furan group of 25a undergoes hydrophobic packing with residues in the EUBP, such as L1313a, F1795a, V232Pb, F2873b, and C3373b. On the cytoplasmic side, the furan ring of 25a forms similar interactions in a symmetric manner (Fig. 6c, e).

a The concentration dependent blockade of urea transport of hUT-A2 by 25a using urease assay. Data are shown as mean ± SEM of three independent experiments (n = 3). b The cryo-EM density (orange) of 25a in hUT-A2 homotrimer from top view or side view. The surfaces of three hUT-A2 monomer are represented by orange, green and blue, respectively. c, d The interactions between hUT-A2 and 25a in extracellular blocker binding pocket (EBBP) (c) or cytoplasmic blocker binding pocket (CBBP) (d). The hUT-A2 are represented in blue cartoon and the key amino acids are represented by blue sticks. The 25a was represented by orange stick-ball. e, f The cutaway view of binding pockets of 25a in extracellular side (e) or cytoplasmic side (f). In the extracellular side, the 25a occupied two subpockets, the EBBP1 (extracellular blocker binding pocket 1) and EUBP (extracellular urea binding pocket), represented by red or blue dashed line respectively. In the intracellular side, the 25a occupied CBBP1 (violet) and CUBP (blue). Residues forming polar interactions or hydrogen bonds with 25a are depicted using triangles. Residues only forming hydrophobic or van der Waals interactions are depicted as solid round circles. The 25a are represented by stick-ball and the hUT-A2 is represented by surface. g Sequence alignment of different UTs in the EBBP1 and CBBP1 regions. The nonconserved amino acids are highlighted with blue background. h The diagram of 25a engaging with the urea binding pocket or potential selective binding pocket with the interfaces represented by blue or violet dashed lines, respectively. Key residues interacting with 25a in these two pockets are shown in blue and violet rectangles, respectively. i, j Effects of hUT-A2 mutants F120L, F120V and L202V on the 25a activity toward hUT-A2 using urease assay. Values are mean ± SEM from three independent experiments (n = 3). The fold changes of IC50 are calculated by dividing the mean value of the mutants by the mean value of the wild type.
Notably, in addition to interacting with residues of the EUBP and CUBP, 25a occupies two additional pockets on the extracellular side and intracellular side, which are named the extracellular blocker binding pocket 1 (EBBP1) and the cytoplasmic blocker binding pocket 1 (CBBP1) (Fig. 6e, f). The EBBP consists of residues from helices 3a and 3b–4b, the LECL, and the ECL2b, and the CBBP consists of residues from helices 3a and 3b, the ICL2a, and the LICL (Supplementary Fig. 9e, f). The benzene ring structure of 25a forms π-π stacking interactions with F1353a of the EBBP on the extracellular side or F1203a, and F3003b of CBBP on the intracellular side (Fig. 6c–f). The terminal end of 25a reached L202LECL, A3264b and P336ECL2b at the pocket end of the EBBP on the extracellular side but reached a relatively open space by contacting L363LICL of CBBP on the intracellular side (Fig. 6c–f). Consistent with these observations, the mutations F120L, F120V, and L202V diminished the binding of hUT-A2 with 25a (Fig. 6g–j and Supplementary Fig. 9g, h).
Collectively, these structural and mutational analyses indicated that the urea channel blocker 25a, by binding to both ends of the transmembrane transport channel (EUBP and CUBP), competes with urea to form hydrogen bonds with the conserved QPb-T5b-T5a-QPa motif necessary for urea transport (Fig. 3f), thereby competitively inhibiting the activity of UT.
Potential of selective UT-A inhibitor design by targeting the EBBP and CBBP
Synthetic compounds with selective activities for UT-A blockade could be used as urea diuretics where conventional salt-transporter blocking diuretics are limited. We next analyzed whether the structure of 25a-hUT-A2 provided clues for selective UT-A inhibitor design. Structural comparison suggested that the amino acids of L202LECL and P336ECL2a of the EBBP in hUT-A2 are replaced by V203LECL and A337ECL2a in hUT-B (Fig. 6g, h), decreasing the hydrophobic volume of the side chains. Molecular dynamics simulation suggests that these changes weaken the hydrophobic interaction between 25a and UT-B (Supplementary Fig. 9i). On the cytoplasmic side, F1203a of the CBBP of hUT-A2 is replaced by L1213a in UT-B (Fig. 6g), which disrupts π-π interactions with the benzene ring of 25a and thus may decrease its binding affinity with UTs. Consistently, the mutations L202V and F120L of hUT-A2 significantly decreased the IC50 of 25a (L202V-IC50 = 3.41 ± 0.01 μM; F120L-IC50 = 1.65 ± 0.02 μM) (Fig. 6i, j). These results indicate that the selective residue differences in the EBBP and CBBP of UT, such as the nonconserved F1203a and L202LECL, could be exploited for selective and potent hUT-A2 inhibitor design (Fig. 6h).
Structural basis of the selective inhibition of UT-A by ATB3
The aminothiazolone derivative UTAinh-B3 (ATB3) is a competitive inhibitor of hUT-A2 (IC50 = 6.38 ± 0.54 μM toward UT-A2), showing approximately 6-fold stronger inhibition potency toward UT-B (Fig. 7a and Supplementary Table 4). To understand the mechanism underlying the selective inhibition of hUT-A2 by ATB3, we solved the cryo-EM structure of ATB3-hUT-A2 at an overall resolution of 3.3 Å (Supplementary Fig. 3a–f). EM densities corresponding to ATB3 were unambiguously detected on both the extracellular and intracellular sides (Figs. 2a, 7b–d and Supplementary Fig. 10a, b). On the extracellular side, the phenyl ring of ATB3 is rotated toward the channel axis by approximately 50°, and the thiazole heterocycle is shifted upward by approximately 1.5 Å compared with 25a (Fig. 7e). This difference in position enables the contact of ATB3 with F2823b in EBBP2 (constituted by F1353a, L202 LECL, F2823b, and L2843b), which does not interact with 25a in the 25a-hUT-A2 complex structure (Fig. 7c, e, f and Supplementary Table 6). Specifically, ATB3 forms both polar and hydrophobic interactions with hUT-A2. In the EUBP, the nitrogen atoms and carbonyl group of the thiazole ring of ATB3 form hydrogen bonds and polar interactions with Q231Pb and T3385b (Fig. 7c, f). Whereas the ortho-methoxy group of ATB3 forms hydrophobic interactions with L2843b and P336ECL2b in EBBP1, the benzene ring and the meta-methoxy group form π-π stacking and hydrophobic interactions with F1353a, L202LECL, F2823b, and L2843b in EBBP2 (Fig. 7c, f). On the cytoplasmic side, the thiazole ring is shifted by 1.5 Å away from the Sm region of the channel and forms polar interactions with Q67Pa and T1763a in the CUBP (Fig. 7d and Supplementary Fig. 10c, d). Notably, the thiazole ring of ATB3 forms extensive polar interactions with the “QPb–TPb–T3a–QPa” motif of the UT channel, which is essential for UT transport. We speculated that competing with urea for interactions with the “QPb–TPb–T3a–QPa” motif underlies the inhibition mechanism of the UT inhibitor ATB3 (Fig. 7c, d and Supplementary Fig. 10d, Supplementary Table 6). Importantly, two specific residues interacting with methoxy groups of ATB3, L202LECL and P336ECL2b, are replaced by V203LECL and A337ECL2b in UTB, respectively (Fig. 7g–i). MD simulation and mutational analysis indicated that these amino acids differences in structurally equivalent residues at the L202LECLP336ECL2b motif (“LP” pocket) contribute to the selective inhibition of hUT-A2 by ATB3 compared with UTB (Fig. 7j–l and Supplementary Fig. 9g, h).

a The concentration dependent blockade of urea transport of hUT-A2 by compound ATB3. Data are shown as mean ± SEM of three independent experiments (n = 3). b The extracellular blocker binding pocket (EBBP) and cytoplasmic blocker binding pocket (CBBP) of ATB3 in hUT-A2. c, d The interactions between hUT-A2 and ATB3 in EBBP (c) or CBBP) (d). The H-bonds are shown as red dashed lines. e Structural comparison of 25a and ATB3 in EBBP. f The ATB3 occupied three subpockets in the extracellular side, the EBBP1, EBBP2, and EUBP. Residues forming polar interactions or hydrogen bonds with ATB3 are depicted using triangles. Residues forming hydrophobic or van der Waals interactions are depicted as solid round circles. The residues F135, L202, and L284 are shared by both EBBP1 and EBBP2. g, h. The methoxy groups of ATB3 formed hydrophobic interaction with L202 and P336 in hUT-A2, whereas ATB3 don’t form similar interactions with structural equivalent amino acid V203 and A337 in hUT-B. The black dashed lines represent the shortest distance. i Barcode comparisons of the residues involving in the EBBP of different UTs. The residues engaged interactions with 25a, ATB3 and HQA2 are colored orange, and the special residues only interact with ATB3 or HQA2 are colored red or green, respectively. The amino acid of hUT-A2 are displayed on the top line and bottom, with the residues in “L-P” pocket and “SCG” pocket colored magenta and green, respectively. The letters inside the black circles represent the non-conserved amino acids of hUT-A3 or hUT-B. j The comparison between the best estimate residue energy contributions of residues L202 or P336 for their interaction with ATB3 and their corresponding L202V, P336A mutants. k Effects of hUT-A2 mutants L202V and P336A on the ATB3 activity toward hUT-A2. Values are mean ± SEM from three independent experiments (n = 3). l The diagram of ATB3 engaging with the urea binding pocket or “L-P” pocket with the interfaces represented by blue or violet dashed lines, respectively.
Structural basis of uncompetitive inhibition of UT-A by CF11
In contrast to the competitive inhibition mode of 25a and ATB3, UTAinh-F11(CF11) was reported as an uncompetitive inhibitor of UT-A, with an EC50 approximately 10 times better for UT-A1 than UT-B (Supplementary Table 4). The IC50 of CF11 toward hUT-A2 was determined to be 2.78 ± 0.32 μM (Fig. 8a and Supplementary Table 4). Importantly, CF11 was observed only on the intracellular side of hUT-A2, without detectable EM density on the extracellular side (Fig. 8b–d and Supplementary Fig. 10e, f). These observations suggested that urea can still bind to the extracellular side of UT-A2 but cannot be transported into the intracellular side; thus, the interaction is unproductive (bound but not released). We therefore speculated that the specific binding mode of CF11 provides a mechanistic explanation for the uncompetitive inhibition of CF11 (Fig. 8b).

a The concentration dependent blockade of urea transport of hUT-A2 by Compound UTAinh-F11 (CF11). Data are shown as mean ± SEM of three independent experiments (n = 3). b The CF11 bound in the cytoplasmic side of hUT-A2 without occupying the urea binding pocket. c The cryo-EM density of CF11 (red mesh) contoured at 4.0 σ level is located in a cavity surrounded by helices 3a–4a, Pa, ICL2a, and LICL. d The key residues of hUT-A2 in CF11 binding pocket are shown by blue sticks. The CF11 is represented by green stick-ball. e The cutaway view of the binding pocket of CF11. The EUBP and the CUPB are represented by blue and green dashed line, respectively. Residues forming polar interactions or hydrogen bonds with CF11 are depicted using triangles. Residues only forming hydrophobic or van der Waals interactions are depicted as solid round circles. f, g Comparison of different amino acids in the CF11 binding pocket between hUT-A2 (f) and hUT-B (g). The A1153a of hUT-A2 is replaced by L1163a of hUT-B, thus cause a steric hindrance to the chlorine atom of CF11. The F1203a of hUT-A2 is replaced by L in hUT-B, therefore losing π–π stacking (orange dashed lines) with the benzene rings of CF11. h Barcode comparisons of the residues involving in the CBBP of different UTs. The residues participated in interactions with 25a, ATB3, and CF11 are displayed as black circles filled orange, and the special residues only interacting with CF11 are shown as black circles filled green. The amino acid sequences of hUT-A2 are shown on top, with the special residues involving in “A-F UCBP” marked by light blue color. The letters inside the black circles filled with light blue represent the non-conserved amino acids in the “A-F UCBP” of hUT-A3 or hUT-B. i The diagram of binding mode of CF11, with the hUT-A2 interfaces represented by blue or light blue dashed lines, respectively. Key residues interacting with CF11 in “A-F UCBP” are shown in blue and light blue rectangles, respectively.
On the intracellular side, CF11 is located in the cavity surrounded by helices 3a–4a, Pa, ICL2a, and LICL (Fig. 8c). CF11 is a γ-sultambenzosulfonamide-type inhibitor, with two benzene rings connected by a nitrogen-sulfur bond. The methoxy group of CF11 inserts into CUBP but does not contact T176, V68, and F341 and only forms hydrophobic contacts with Q67Pa and F70Pa, leaving an empty space in the UT transport channel (Fig. 8d, e and Supplementary Table 7). Therefore, CF11 cannot compete with urea molecules to form polar interactions with the Q231Pb–T3385b–T1765a–Q67Pa motif of the CUBP, a common feature shared by competitive inhibitors 25a and ATB3. These structural features provided additional insights into the uncompetitive inhibition mode of CF11.
The specific residues encompassing the uncompetitive inhibitor CF11 were named the uncompetitive binding pocket (UCBP) (Fig. 8e). Inside the UCBP, CF11 formed hydrophobic interactions with A1153a, M2993b, L363LICL, and V366LICL. The two benzene rings of CF11 form π–π stacking interactions with F1203a, whereas the nitrogen-sulfur atoms connecting the two benzene rings form hydrogen bonds with the hydroxyl group of Y1233a and the main chain atoms of V1745a–F1755a. In addition, CF11 participates in π-π stacking with F3003b (Fig. 8d, e). According to the sequence alignment of different UTs, we found that two nonconserved amino acids in UCBP, A1153a, and F1203a of hUT-A2, may contribute to the selectivity of F11 (Fig. 8f–h). Notably, A1153a in UT-A2 is replaced by L1163a in UT-B, which may provide steric hindrance to the chlorine atom of CF11, decreasing the binding affinity (Fig. 8f, g). This speculation was supported by structure-function relationship studies: replacing the chlorine atom with a hydrogen atom in CF11 improved the inhibition potency against UT-B20. Moreover, F1203a of hUT-A2 is replaced by L in hUT-A3 and hUT-B, losing π-π stacking with the benzene rings of CF11 (Fig. 8f, g). Therefore, A1153a and F1203a of UCBP are key residues of hUT-A2 that contribute to the selectivity of CF11, and we named the pocket the “A-F UCBP”. In summary, specific UCBP residues of UT-A2, such as A1153a and F1203a, could be utilized for inhibitor design targeting hUT-A2 for both uncompetitive inhibition and selectivity (Fig. 8i).
Mechanism of noncompetitive inhibition of UT-A by targeting an “SCG” pocket
The 8-hydroxyquinoline derivative UTAinh-A2 (HQA2) is a noncompetitive inhibitor of UT-A, with selectivity against UT-B of more than tenfold (IC50 for UT-A1 vs. UT-B is approximately 5 μM vs. 50 μM) (Supplementary Table 4)21. The IC50 values of HQA2 for blocking urea transport mediated by hUT-A2 were 3.66 ± 0.09 μM (Fig. 9a). In the HQA2-hUT-A2 complex structure, only one binding site located on the extracellular side of hUT-A2, located in the cavity surrounded by helices 3a, 3b–4b, and Pb, and the LECL, and ECL2b (Fig. 9b, c). Distinct from all other competitive inhibitors and uncompetitive inhibitors, which bind perpendicularly to the extracellular side of UT, HQA2 binds parallel to hUT-A2. Notably, the benzene ring of HQA2 is located in EBBP1 and EBBP2 (EBBP1-2). In EBBP1-2, the benzene ring of HQA2 forms π–π stacking with F1353a and hydrophobic interactions with L202LECL, L2843b, P335ECL2b, and P336ECL2b, including interactions with the “L-P” selective pocket (Fig. 9d, e and Supplementary Table 8). In particular, the methoxy group in the benzene ring of HQA2 extends into the EUBP, forming a hydrogen bond with the side chain of Q2315b (Fig. 9d, e), without occupying the entire EUBP or the CUBP, thereby affording a potential mechanism of noncompetitive inhibition of urea transport.
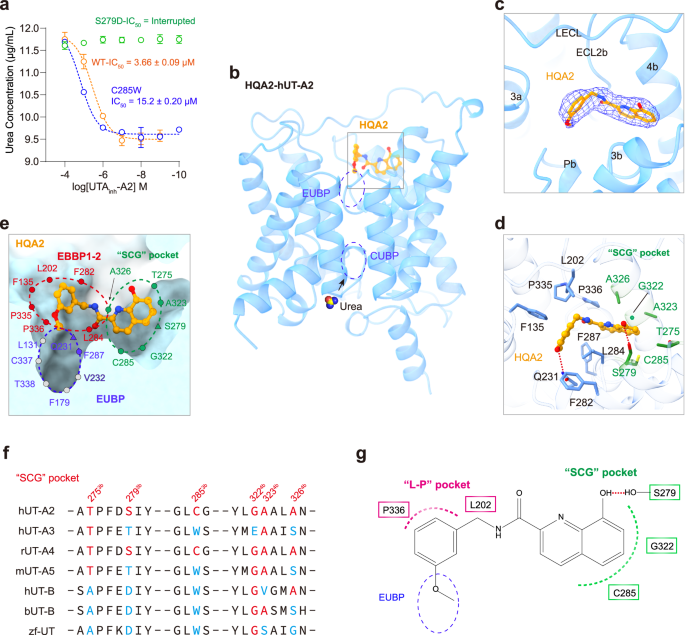
a The concentration dependent blockade of urea transport of wild type hUT-A2 and S279D, C285W mutants by HQA2. Values are mean ± SEM from three independent experiments (n = 3). b The HQA2 bound in the extracellular side of hUT-A2 without occupying the urea binding pocket. The hUT-A2 are represented in blue cartoon and the HQA2 shows as orange stick-ball. c The cryo-EM density (blue mesh) of HQA2 at the 4.0 σ contour level is located in the cavity surrounded by helices 3a, 3b–4b, and Pb, and the LECL, and ECL2b. d The key residues of the HQA2 binding pocket of hUT-A2 are represented by sticks and the residues involving in the “SCG” pocket are colored green. The H-bond are shown as red dashed line. e The cutaway view of the binding pocket of HQA2 (orange stick-ball). The EUBP, EBBP1-2 and the noncompetitive “SCG” pocket are represented by blue, red and green dashed line respectively. Residues forming polar interactions or hydrogen bonds with HQA2 are depicted using triangles. Residues only forming hydrophobic or van der Waals interactions are depicted as solid round circles. The HQA2 are represented by orange stick-ball and the hUT-A2 are represented by surface. f Sequence alignment of the structurally equivalent residues in the noncompetitive SCG pocket of hUT-A2 compared with other UT members including hUT-A3, rUT-A4 (rat UT-A4), mUT-A5 (mouse UT-A5), hUT-B, bUT-B (bovine UT-B) and zf-UT. Key residues of SCG pocket in hUT-A2 are shown in red font on the top line. Sequences with the same amino acids as the SCG pocket of hUT-A2 are represented in red, while those with non-conserved amino acids are represented in light blue. g The diagram of HQA2 engaging with the extracellular urea binding pocket (EUBP), L-P pocket and the noncompetitive SCG pocket with the interfaces represented by blue, violet and green dashed lines, respectively. Key residues interacting with L-P pocket and SCG pocket are shown in violet and green rectangles, respectively. The H-bond between HQA2 and residues S279 is shown as red dashed line.
Importantly, the hydroxyquinoline moiety is rotated by approximately 90° and occupies a side pocket encompassing 6 residues with small side chains, named the SCG pocket (Supplementary Fig. 10g). In the SCG pocket, the hydroxyl group of the hydroxyquinoline moiety forms a hydrogen bond with the side chain of S2793b and van der Waals interactions with T275ECL1b, C2853b, G3224b, and A3234b (Fig. 9d and Supplementary Fig. 10h, i). Sequence alignment showed that the amino acid sequences in the SCG pockets of different UTs are not conserved, especially for residues S2793b, C2853b, and G3224b (Figs. 7i, 9f). Importantly, mutations of SCG pocket residues to structural equivalent residues in other UT members, such as S279D or C285W, significantly decreased the activity of HQA2 in blocking urea transportation (Fig. 9a and Supplementary Fig. 7k, l). These results suggested that this pocket has potential for designing hUT-A/B selective inhibitors. Moreover, the region where the SCG pocket is located is completely independent of the urea transport channel of hUT-A2 and may therefore serve as an allosteric pocket for ligand binding to regulate UT-mediated urea transportation (Fig. 9e–g).