Bacterial strains used and their maintenance
All Bifidobacterial strains were obtained from the German culture collection center (DSMZ, Germany). They were grown in De Man, Rogosa, and Sharpe (MRS) Brain Heart Infusion (BHI) or Nutrient Broth Media (NB). Various anti-tubercular drugs, such as rifamycin (RIF), streptomycin (Sm), isoniazid (INH), and pyrazinamide (PYR), were obtained from Hi-Media Laboratories (Mumbai, India). B. adolescentis being microaerophilic, at the initial stages of growth, they were first streaked from the glycerol stock to a MRS plate, and they were then incubated in a small anaerobic jar apparatus (with a gas pack containing 3.5 L of polycarbonate, Hi-Media Laboratories, Mumbai, India) at 37 °C for 72 h. After 72 h, small isolated colonies were inoculated into fresh MRS broth in a 15 mL Falcon tube with 10–12 mL of media incubated at 37 °C without shakinguntil an OD 600 of 2.0 was reached. Glycerol stocks were made with these cultures and stored at -80 °C.
Growth curve studies
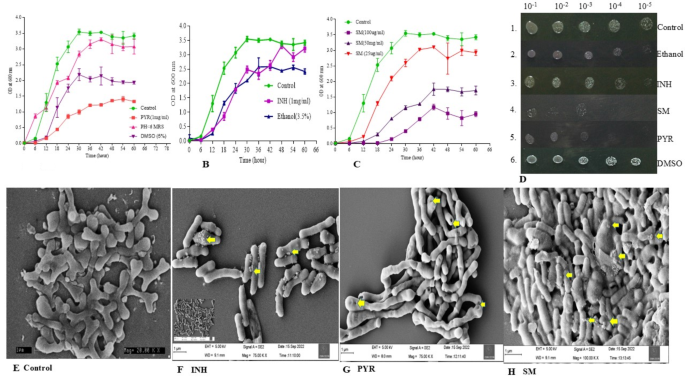
The growth curves of the various Bifidobacteria, such as B. adolescentis and B. asteroids, were studied at 37 °C. The following protocol was strictly followed: A total of 10 mL of freshly prepared MRS broth was cultured at 37 °C without agitation. Then, fresh streaking was impeccably done and single-isolated colonies were introduced into it. Next, 4.0% of the freshly grown cultures were subcultured into 50 mL of MRS medium to start the growth curve at a plate number of 0.01–0.05. The growth curve was closely monitored under the same circumstances. Bacterial samples were periodically taken every 12 h and absorbance was calculated using a precise plate reader scale.
Preparation of anti-tubercular drugs
Three anti-tubercular drugs were considered for the growth curve studies: PYR, INH, and Sm. The miscibility of each drug differs from each other. PYR is dissolved in DMSO (5.0%), INH is soluble in 3.5% ethanol, and SM is soluble in water. Therefore, after dissolving the drug, the growth curves were followed, wherein only DMSO and ethanol were considered as the right controls. In the case of PYR growth curve studies, two controls were considered: (1) a low pH, and (2) 5.0% DMSO.
Analysis of growth pattern in the presence and absence of anti-tubercular drugs
4.0% of the Bifidobacteria cultures were promptly transferred to fresh MRS medium (50 mL), which contained various anti-tubercular drugs. We measured the absorbance at OD600 and collected samples every 6 h to analyse the growth curves with and without anti-tubercular medications. We performed each experiment three times, meticulously measuring the absorbance and plotting it against time.
Spot assay
As explained above, 72-hour-grown Bifidobacteria of OD600 1.0 were considered for spot assay. MRS agar plates with and without antibiotics (MIC) were prepared. Subsequently, the resulting 1.0 OD600 cells were serially diluted to 10−5 in fresh MRS media (900 µL media and 100 µL culture). Serial dilutions were made, and one µL of each dilution was spotted on plates with and without antibiotics. Plates were incubated at 37 °C in an anaerobic gas chamber for 48 h and analysed for growth53.
Anti-tubercular drug uptake and surface analysis
We followed standard protocols for measuring anti-tubercular drug uptake, such as HPLC. Since this is a very sophisticated and sensitive, it also helps in the estimation of the approximate molecular weight of a substance54. Once cells have been exposed to anti-tubercular drugs to understand the surface texture and distinguish rough and smooth strains of Bifidobacteria. The agglutination reactions with acriflavine solution (1/1000) and the capacity to uptake crystal violet55 were followed. The treated cells were photographed to understand their morphologies.
B. adolescentis adaptability studies
B. adolescentis (0.5–0.8 OD600) was grown in the presence of PYR (1.0 mg/mL), INH (1.0 mg/mL), and Sm (25–100 µg/mL). Every 6.0 h, samples were withdrawn and harvested by centrifugation at 5000 rpm for 20 min. Subsequently, cells were processed by following Bhat et al.‘s procedure.
To summarize the process, the cells were collected and washed thoroughly. Next, they were dissolved in a potent organic mixture of methanol, chloroform, and water (12:5:3, 3.0 mL) and subjected to a high temperature treatment of 65 °C for 20 min. The cells were then stored at -20 °C for 12 to 16 h, followed by centrifugation. The unlysed cell debris was promptly removed, and the supernatant was collected and centrifuged at high speed for 15 min. The supernatant was then efficiently dried under a vacuum and the pellet was dispersed in 50 mM, 300 µL of phosphate buffer at pH 7.0. With another round of centrifugation, the organic layer was dried, and the samples were reconstituted in an HPLC buffer on the day of analysis, making them ideally suited for HPLC analysis.
Field emission scanning electron microscopy (FE-SEM) studies
The investigation employed Field Emission Scanning Electron Microscopy (FE-SEM) to thoroughly examine the surface morphology of Bifidobacteria that were treated and untreated with anti-tubercular drugs. The procedures for cultivating and collecting the cells are clearly outlined in the growth curve investigation section. After two phosphate buffer washes, the cell pellets were treated with 2.0% glutaraldehyde and incubated at 4.0 °C for 12–14 h. A gradient of 10–100% ethanol was used to wash the cells, and the sample was ultimately resuspended in absolute alcohol (50–100 µL). An aliquot (~ 2.0 µL) was placed on a cover slip, dried, and examined under an FE-SEM. Only visually acceptable and compelling photographs magnified by 20,000 times were taken into consideration for further analysis and presentation.
Genomic DNA isolation to understand the modifications at genome level
To extract genomic DNA, GeneJet Genomic DNA Isolation Kit was used. This kit is manufactured by Fermentas, Inc. 830 Harrington Court, Burlington, ON, Canada. The process involved growing a single isolated colony in an anaerobic gas chamber at 37 °C, followed by inoculation into ten millilitres of fresh MRS broth. After that, lysozyme, an enzyme that lyses cell walls, was added to the cells and heated at 37 °C in a Tris-Cl and EDTA buffer with pH 8.0. Later, cells were treated with proteinase K, RNase A, lysis buffer and incubated at room temperature for 10 min. Later, they were centrifuged at 12,000 rpm for 20 min, and the supernatant was loaded into the GeneJet column. The column was washed with wash buffer, and finally, DNA was eluted with Tris. EDTA pH 8.0 before considering further studies, gDNA was electrophoresed to check the quality.
16s rRNA, rpsL, rrs, and gidB gene amplification
The gDNA purified as described above was used as the template for 16 S rRNA amplification using the primers as stated in Table 1. The PCR parameters were: initial denaturation at 95 °C for 5.0 min, followed by 35 cycles of 95 °C for 30s, 56 °C for 45s, and 72 °C for 1.0 min, and a final extension at 72 °C for 5.0 min. DyNAzime II DNA polymerase (Thermo Scientific) was used for the amplification. After completion of PCR, the DNA was analysed through 1.0–1.5% agarose electrophoresis (based on the molecular weight of the amplified fragment). The same template DNA was used for 16 S rRNA, rpsL, rrs, and gidB amplifications.
Particle size analysis to determine bacterial size
As per the previous reports, light scattering technology may be one of the best, cheaper, and most reproducible ways to understand cell diameter, size, and shape. Light-scattering measurements have a crucial advantage over other methods as they require only a minute quantity of sample. Dilute samples are preferred as they aid in simplifying data processing, minimizing multiple scattering events. For the first time, Wyatt et al. reported the use of light scattering technology to identify bacterial cells56. Therefore, we followed recent technologies that have been extensively used to understand and measure cell shape and diameter. The procedure begins with the growth of Bifidobacteria in MRS for 72 h at 37 °C in the presence and absence of PYR, INH, and Sm. Subsequently, harvested cells were washed twice with phosphate buffer and re-suspended in fresh phosphate buffer, vortexed to remove the cell clumps, and used for the scattering study. Particle size has been analysed by a Microtrack particle size analyzer (Microtrac SDC; 90–250 VAC; 47–63 Hz, USA). Corresponding chromatograms were collected and analysed.
Catalase foam assay
In Tadayuki Iwase et al. (2013), a simple assay to measure catalase activity was followed. The assay is, in brief, A Simple Assay for Measuring Catalase Activity57. We followed this methodology because of its simplicity and because it is a qualitative approach for measuring catalase activity. This study employs commonly available chemicals such as hydrogen peroxide, Triton X-100, and commercial catalase.
Various + ve and -ve controls for the assay, such as Salmonella typhi (+ ve, grown in BHI media) and Lactococcus lactis (-ve), were considered. B. adolescentis, along with the controls, were grown in fresh MRS media (10 mL). Overnight grown 0.5 and 1.5 OD600 cells (approximately 105–107 cells) of 2.0 mL each in transparent test tubes were harvested and washed with phosphate buffer twice. Later, centrifuged to obtain a pellet, 100 µL of Triton X-100 and 100 µL of H2O2 (30%) were sequentially added. The reaction mixtures were mixed well and incubated at room temperature for 5–10 min. Upon completion of the reaction, the height of the O2-forming foam in the reaction tube was measured with a scale. The various tubes with various samples of O2 foam were photographed. The foam assay kinetics were followed, wherein samples of bacterial cultures were withdrawn at 0, 6, 12, 18, 24, 30, and 36 h. Each sample was processed as explained. The graph was drawn between time and the height of the foam formed, and pictures were taken accordingly.
Catalase gel assay / Zymogram
An established catalase zymogram assay procedure by Magdalena et al. (2018) was employed58. Briefly, the process is as follows: The native gels were electrophoretically separated, washed in MQ for ten minutes, incubated in a solution of freshly prepared 0.01% OR 4.0 mM H2O2 (made from a 30% stock) for ten minutes, and then rinsed in 100 mL of MQ for thirty minutes. The gels were then incubated in a freshly prepared solution containing 1.0% ferric chloride hexahydrate and 1.0% potassium ferricyanide trihydrate while being gently agitated. One can observe the formation of a set of strong achromatic bands on a green-blue background. To avoid overstaining, immediately the gel was removed from the staining solution and placed in MQ water.
Growth curve studies of B. adolescentis in presence and absence of H2O2
To understand the ability of B. adolescentis to sustain physiological concentrations of H2O2, we followed the growth curve in the presence of various concentrations of H2O2, such as 1.4 and 6.0 mM. The procedure followed in brief: a fresh streaked plate containing a single isolated colony was inoculated into 10 mL of fresh MRS and was grown at 37 °C without shaking under microaerophilic conditions as stated above. Subsequently, after 72 h of growth, a 1.0% culture was inoculated into fresh MRS broth in the presence of two different concentrations of H2O2, as stated above. The OD600 at the beginning of the growth curve studies was kept at 0.05; samples were collected at various intervals, and growth was followed for 60 h. Finally, the graph was plotted between OD600 and time.
MIC determination
The micro-broth dilution method was followed to determine the MIC of B. adolescentis to anti-tubercular drugs such as PYR, INH, and SM59. The method followed in brief: B. adolescentis was grown as explained above. Once the absorbance value of the cells hit 0.4, they were categorized as being in the mid-exponential growth phase. Following this, the culture was promptly adjusted to a final cell count of 4.0 × 105 CFU/mL to ensure optimal growth and development. 100 µL of bacterial cells were introduced into each well. Subsequently, the highest to the lowest concentration of anti-tubercular drugs were added, making the final volume of the reaction 200 µL. Immediately, serial dilutions were followed that brought down the drug concentration. The reaction mix was mixed well and incubated at 37 °C overnight without shaking. The minimum inhibitory concentration (MIC) was unequivocally determined by the observation of no discernible growth within the well. The MIC was followed for PYR, INH, and Sm for Bifidobacteria three times.
Particle size analysis
After being cultivated in MRS broth for 36 h, B. adolescentis was sub-cultured at a 1.0% inoculum level into 15 ml of MRS broth supplemented with various antitubercular drugs. Such as 25 µg/mL, 50 µg/mL, and 100 µg/ml of streptomycin, as well as Pyrazinamide (PYR) dissolved in 100% dimethyl sulfoxide (DMSO) and isoniazid (INH) dissolved in 70% ethanol. In addition, PYR-treated cultures were exposed to low pH levels using acidic MRS media; a control culture that was just grown MRS broth was also included. In order to obtain cell pellets, cultures were centrifuged at 6000 rpm for 15 min using a REMI R-24 centrifuge, after being cultured under microaerophilic conditions for 36 h at 37 °C. Pellets were re-suspended in 1.0 mL of double-distilled water for analysis after being cleaned in autoclaved phosphate-buffered saline (PBS) at pH 7.0. A Microtrac particle size analyzer was used to evaluate the distribution of particle sizes (MICROTRAC SDC; 90-250VAC; 47–63 Hz, USA). Dynamic Light scattering (DLS) was an institutional facility that analyzes particle size ranging 0.8–6500 nm.
Whole genome sequence analysis and AMR prediction
We have employed a variety of bioinformatics tools and databases, including the Comprehensive Antibiotic Resistance Database (CARD), to comprehend the resistance of Bifidobacteria adolescentis. CARD60 was created to record gene sequences carrying multiple antibiotic resistance genes together with pertinent metadata. Resistance gene identifier (RGI) and BLAST are further CARD tools for quick identification and visualisation of antibiotic resistance genes in new, unannotated genomes. Information gathered from systematic reviews for N. gonorrhoeae AMR was incorporated in a recent release (3.0.3) of CARD. Kubanov et al. used the RGI web portal from CARD to search for and analyse AMR genetic determinants in genomic sequences of N. gonorrhoeae strains61,62,63.
Rapid annotation using subsystems technology (RAST)
RAST is another tool used to predict AMR profiles in a given bacterium. B. adolescentis whole genome sequence was analysed for the presence of various AMR genes by using RAST. We were mainly interested in looking for the antibiotic-resistance pathways present in the genome.
CARD (comprehensive antibiotic resistance database) data analysis
As said above, this is a tool used to identify AMR genes, AMR gene family, Drug class, and resistant mechanism existing.
Determination of important virulence, disease, and defence related genes
The genome of Bifidobacterium was functionally annotated with the NCBI prokaryotic genome annotation pipeline (PGAP) v. 4.7 and rapid annotations using subsystems technology (RAST)60,62. Figure 4A–F show the RAST annotation antibiotic resistance pathway present in Bifidobacteria. Further, CARD data analysis shows AMR genes, gene families, drug classes, and the resistant mechanisms involved.
Using the genome functional annotations, the presence of virulence, disease, and defence genes was manually searched and discovered in 54 different categories. Within the category of antibiotic and hazardous chemical resistance, there were thirty-one subcategories. They include beta-lactamase (1), ribosome protection types to (5), tetracycline resistance, bile salt hydrolysis (2), cobalt-zinc-cadmium resistance (5), tetracycline resistance, ribosome protection type (5), resistance to fluoroquinolones (6), and multidrug resistance efflux pumps (2).
3D-structure prediction/homology modelling and docking studies
We utilized the canonical amino acid sequence from NCBI (https://www.ncbi.nlm.nih.gov) to determine the homology model of Nicotinamidase/pyrazinamidase, the S12 protein, and its mutations in B. adolescentis. We determined the Nicotinamidase/Pyrazinamidase S12 protein structure and mutations using the I-TASSER suite (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). In the PDB database, we found that nicotinamidase/pyrazinamidase and its mutations were unequivocally similar to 3PL1 (Pyrazinamidase of Mtb), whereas s12 protein and its mutants were unmistakably similar to 7BGD (s12 protein of S. aureus). The model with the greatest confidence score was utilized for the structural evaluation and docking investigations. Pyrazinamide and streptomycin, two anti-tubercular drugs, were unequivocally used as ligands for their corresponding proteins. Docking was performed in AutoDock Tools (ver. 1.5.7) and the complex was visualized and processed using Discovery Studio 2020.