Biochemists from the National University of Singapore (NUS) have discovered a new subclass of trifunctional enzymes in gram-positive bacteria, which play a critical role in the biosynthesis of an antimicrobial lanthipeptide.
Lanthipeptides are a group of ribosomally synthesized and post-translationally modified peptides (RiPPs) synthesized by gram-positive bacteria in response to environmental stress. They are important biomolecules with functions including antibacterial, antifungal, and antiviral effects and hold promising potential for developing new drugs.
Despite their therapeutic significance, the mechanism underlying the post-translational modification of a particularly robust class of these peptides remains largely unexplored.
Using a combination of bioinformatics, cryo-electron microscopy (cryoEM) structural characterization, and functional assays, the research team led by Assistant Professor Luo Min from the Department of Biological Sciences at NUS characterized a distinct subclass of lanthipeptide modification enzymes.
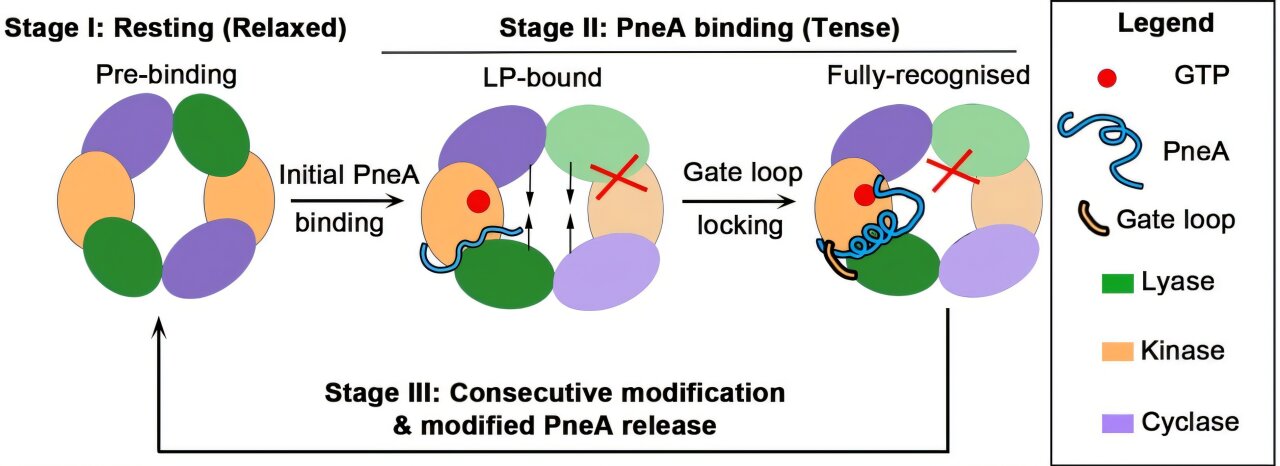
These enzymes are responsible for recognizing the precursor lanthipeptide and sequentially installing a series of post-translational modifications, changing the peptide into its functional conformation.
In particular, they reported the first homodimeric lanthipeptide modification enzyme, PneKC from the bacteria species Streptococcus Pneumonae. Structural analysis of the enzyme revealed a critical region, or “dimerization hotspot,” where two parts of the enzyme join to form a dimer. When this region was mutated, the enzyme lost its ability to form this dimeric structure.
The findings were published in the journal Nature Communications.
Functional assays showed that the enzyme dimer self-regulates through a mechanism called negative cooperativity. In this process, when a substrate binds to one-half (protomer) of the dimer, it triggers a change in its shape. This change is transmitted across the dimerization interface to the unbound protomer, effectively controlling its activity.
The team also uncovered the initial stages of peptide recognition and proposed a set of previously unidentified catalytic residues, which are specific amino acids within the enzyme’s active site that directly facilitate the final step of peptide modification. These catalytic residues play a crucial role in driving the cyclization step, a key step that enables the peptide to mature into its active form as a lanthipeptide.
This step had long remained a mystery in this class of enzymes, and the identification of these catalytic residues provides a critical piece of the puzzle, enhancing our understanding of how these enzymes facilitate the complex modifications essential to lanthipeptide biosynthesis.
Following this discovery, Ms. Yifan Li, a Ph.D. candidate on the research team and the first author of this paper, is now focused on uncovering the next stages of lanthipeptide modification. Her current work aims to understand the subsequent maturation processes and pharmacodynamics of these newly identified lanthipeptides, with the goal of advancing their potential therapeutic applications.
Prof Luo said, “Our current study significantly enhanced our understanding of the dynamic processes underlying peptide recognition and domain coordination within lanthipeptide modification enzymes. Moreover, considering the high potential of lanthipeptides in antimicrobial applications, these insights pave the way for the development of lanthipeptides for antimicrobial purposes.”
More information:
Yifan Li et al, Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers, Nature Communications (2024). DOI: 10.1038/s41467-024-51600-6
Provided by
National University of Singapore
Citation:
Biochemists uncover a new enzyme subclass in antimicrobial lanthipeptide biosynthesis (2024, November 29)
retrieved 30 November 2024
from https://phys.org/news/2024-11-biochemists-uncover-enzyme-subclass-antimicrobial.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.