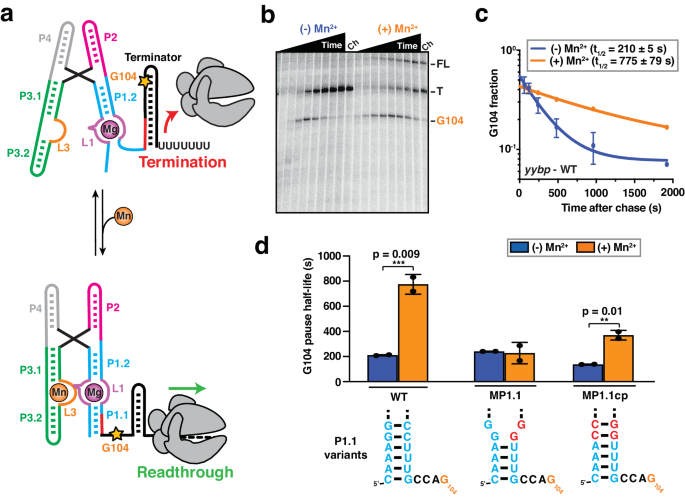
Ligand-induced stabilization of riboswitch helix P1.1 modulates RNAP pausing efficiency
Transcriptional pausing has been shown to be an inherent parameter that enables co-transcriptional RNA folding and protein recruitment to play an active role in regulating gene expression27,35. Therefore, we evaluated how riboswitch folding modulates transcriptional pausing, using single-round transcription assays (Fig. 1b). We identified a long-lived intrinsic (elemental) pause site at position G104 in the yybP expression platform, which is four nucleotides downstream of the P1.1 switch helix (Fig. 1b and Supplementary Fig. 1) and closely related to the known bacterial consensus pause sequence in the DNA template34. Interestingly, the duration of the G104 pause was significantly extended in the presence of saturating Mn2+ ions (Fig. 1c and Supplementary Table 2), indicating that the ligand-bound state further promotes RNAP pausing at this position. Control experiments performed with a mutant abolishing Mn2+ binding to the riboswitch (A41U)13 showed no such difference, supporting that promotion of RNAP pausing is the result of riboswitch folding in the ligand-bound state, rather than a direct effect on the enzyme (Supplementary Table 2).
Upon examination of the riboswitch secondary structure relative to the G104 pause position in the RNAP active site, we hypothesized that the formation of the P1.1 switch helix inside the RNAP exit channel stabilizes transcriptional pausing (Supplementary Fig. 1). To test its involvement in RNAP pausing, we destabilized the P1.1 helix by mutating residues C95 and C96 to G95 and G96, respectively (mutant MP1.1; Fig. 1d and Supplementary Table 2). Consistent with our hypothesis, MP1.1 completely abolishes the impact of Mn2+ on RNAP pausing efficiency. Importantly, compensatory second-site mutations G5C and G6C (mutant MP1.1cp) not only restore two base pairs at the top of P1.1 but also the Mn2+-induced enhancement of RNAP pausing to levels similar to those of the wild-type (Fig. 1d and Supplementary Table 2).
Overall, these data are consistent with a regulatory system in which the ligand-mediated stabilization of RNA helix P1.1 in the RNAP exit channel extends transcriptional pausing.
Ligand-mediated folding of the P1.1 helix depends on the proximity of RNAP
To characterize the co-transcriptional folding of the riboswitch P1.1 helix as a function of ligand binding, we used an in vitro transcription assay that generates a co-transcriptionally folded PEC with a single fluorophore at the 5â end for identification and subsequent probing at the single-molecule level (Fig. 2a, b)22. We added a short (8-nucleotide), Cy5-labeled DNA SiM-KARTS (Single-Molecule Kinetic Analysis of RNA Transient Structure) probe against the 5â segment of the P1.1 helix, which allowed for dynamic probing of solvent accessibility of the C2-G8 switching region (Fig. 2b and Supplementary Fig. 1)23,53,54. Upon simultaneous excitation of Cy3 and Cy5, single-molecules were located on the microscope slide through their Cy3 (green) PEC-104 fluorescence signal, whereas the accessibility of the target switching region over time was detected through spikes of the corresponding SiM-KARTS fluorescence intensity in the Cy5 (red) channel (Fig. 2c, d). The cumulative dwell time distributions of the SiM-KARTS probe in the 5â P1.1 unbound (Ïunbound) and bound (Ïbound) states were fitted with double-exponential functions to extract the corresponding binding (kSK-on) and dissociation (kSK-off) rate constants, respectively (Fig. 2e, f).
a Nascent fluorescently labeled Paused Elongation Complexes (PEC) are transcribed in vitro using E. coli RNAP. The halted complex (EC-13) is prepared through the addition of a dinucleotide labeled with Cy3 (ApU-Cy3) and UTP deprivation (ATP/CTP/GTP) to halt the RNAP at the end of the U-less ITR. The biotin-streptavidin interaction at the 3â-end of the DNA template constitutes a stable transcriptional roadblock to stall the RNAP at the desired position. b SiM-KARTS experimental setup. Immobilization of the PEC on the microscope slide occurs through the biotin-streptavidin roadblock. Repeated bindings of the SiM-KARTS probe targeting the 5â segment of P1.1 are monitored through direct excitation of the Cy5 fluorescent dye. Representative single-molecule trajectories showing the SiM-KARTS probe binding (pink) to 5â segment of the P1.1 helix of PEC-104 recorded in the absence (c) or presence (d) of 0.5âmM Mn2+ added co-transcriptionally. Hidden Markov modeling (HMM) is indicated on the top of each trace. A.U., arbitrary unit. Plots displaying the cumulative unbound (e) and bound (f) dwell times of the SiM-KARTS probe in the absence (blue) and presence (orange) of 0.5âmM Mn2+ in the context of PEC-104. The binding (kon) and dissociation (koff) rate constants of the SiM-KARTS probe are indicated. The reported errors are the error of the fit. Total number of molecules analyzed for each condition is as follow: (â) Mn2+â=â350; (+) Mn2+â=â246. g Overall binding rate constants (kon) of the SiM-KARTS probe in the context of PEC-104 WT, MP1.1 and RT-96 constructs determined in the absence (blue) and presence (orange) of 0.5âmM Mn2+. Error bars are the SD of the mean from independent replicates. The statistical significance of differences was determined using the two-tailed Studentâs t-test (***pâ<â0.01, **pâ<â0.05, *pâ<â0.1). See also Supplementary Fig. 2. Total number of molecules analyzed for each condition is as follow: PEC-104 WT (â) Mn2+â=â350; PEC-104 WT (+) Mn2+â=â246; PEC-104 MP1.1 (â) Mn2+â=â330; PEC-104 MP1.1 (+) Mn2+â=â292; RT-96 (â) Mn2+â=â118; RT-96 (+) Mn2+â=â182. Source data are provided with this paper at https://doi.org/10.7302/22513.
In the presence of near-physiological 1âmM Mg2+ only, the SiM-KARTS probe bound to its 5â P1.1 target with two rate constants of 3.67â±â0.02 Ãâ106 Mâ1 sâ1 (kSK-on,fast; fraction 59%) and 0.63â±â0.01 à 106 Mâ1 sâ1 (kSK-on,slow; fraction 41%), as well as dissociated with two rate constants of 0.55â±â0.01âsâ1 (kSK-off,fast; fraction 61%) and 0.08â±â0.01âsâ1 (kSK-off,slow; fraction 39%). This observation is consistent with the notion that the SiM-KARTS probe senses (at least) two alternative structures of the riboswitch P1.1 helix that could arise on the same RNA molecule. Indeed, the targeted C2-G8 region is expected to either form the P1.1 helix or adopt a single-stranded conformation, which the absence of Mn2+ ligand should favor12,13.
To test the latter notion, we next transcribed PEC-104 in the presence of a saturating concentration (0.5âmM) of Mn2+ (generally added together with 0.5âmM Mg2+ for a consistent total concentration of 1âmM divalents) and again probed the 5â P1.1 segment using SiM-KARTS (Fig. 2d). Under these conditions, the probe binding rate constants were 2.93â±â0.01 à 106âMâ1 sâ1 (kSK-on,fast; fraction 44%) and 0.49â±â0.01 à 106 Mâ1 sâ1 (kSK-on,slow; fraction 56%), whereas the two dissociation rate constants were 0.36â±â0.01âsâ1 (kSK-off,fast; fraction 85%) and 0.08â±â0.01âsâ1 (kSK-off,slow; fraction 15%). Notably, both the faster and slower binding rate constants decreased by ~20% compared to the no-ligand condition (Fig. 2e). In addition, the fractional contribution of kSK-on,slow increased (from 41% to 56%) when co-transcriptionally adding Mn2+, at the expense of the kSK-on,fast fraction. Consequently, the weighted average binding rate constant (kSK-onoverall) decreased by ~37% (from 2.42â±â0.17 à 106 Mâ1 sâ1 to 1.53â±â0.15 à 106 Mâ1 sâ1) upon addition of Mn2+ ions (Fig. 2e and Supplementary Table 3). These observations support the notion that ligand binding to the riboswitch in the context of PEC-104 promotes P1.1 helix formation and thus limits SiM-KARTS probe access, albeit not completely. Notably, binding of the SiM-KARTS probe effectively mimics formation of helix P1.1, albeit as an RNA:DNA hybrid (Fig. 2b). The observed slight decrease in overall SiM-KARTS probe dissociation rate constant kSK-offoverall (from 0.37âsâ1 to 0.32âsâ1; Fig. 2f and Supplementary Table 3) upon addition of Mn2+ is therefore also consistent with the ligand stabilizing formation of the P1.1 helix. As a control experiment, we performed SiM-KARTS probing of a PEC-104 with our mutant MP1.1 in which nucleotides C95 and C96 were both mutated to G, disrupting the upstream Watson-Crick base pairs of the P1.1 helix (Supplementary Fig. 1). While the overall SiM-KARTS binding rate constant expectedly decreased, there was still a similar decrease (35%) associated with the addition of Mn2+ (Fig. 2g, Supplementary Fig. 2 and Supplementary Table 3), suggesting that the ligand influences the entire P1.1 helix surveyed by the SiM-KARTS probe.
To ask whether the proximal RNAP of PEC-104 participates in the ligand-dependent modulation of the P1.1 helix, we next performed SiM-KARTS probing on a co-transcriptionally folded transcript from which the RNAP was run off by omission of the 3â biotin-streptavidin block. To this end, transcription was performed on an extended DNA template, installing an additional 3â end RNA sequence as âanchorâ that was hybridized with a 5â biotinylated capture probe (CP) for subsequent immobilization of the transcript on the microscope slide23. Importantly, this released transcript (RT-96) comprised the full yybp riboswitch up to residue C96 to allow for formation of the upper base pairs of the P1.1 helix, mimicking the entire RNA transcript upstream of the RNA-DNA hybrid in the PEC. Strikingly, no effect of Mn2+ on the RT-96 riboswitch was detected by the SiM-KARTS probe, yielding comparable rate constants in the presence and absence of ligand that match the kinetic parameters for PEC-104 in the absence of ligand, where the P1.1 helix is not (fully) formed (Fig. 2g, Supplementary Fig. 2 and Supplementary Table 3). These observations indicate that the presence of RNAP in PEC-104 actively assists P1.1 helix folding upon addition of Mn2+.
Taken together, our single-molecule probing reveals that ligand and RNAP, paused at a specific downstream site, collaborate to nucleate the P1.1 switch helix. Put differently, the riboswitch integrates disparate signals from distal Mn2+ binding and proximal RNAP pausing for gene regulatory action.
The presence of RNAP promotes the docked conformation of the riboswitch
Our biochemical and single-molecule probing assays underscore the importance of both RNAP and ligand for riboswitch folding. However, SiM-KARTS only reveals structural rearrangements that occur locally (P1.1 stem) and does not report the intramolecular dynamics for a single RNA molecule. Generally, the presence of RNAP and DNA template during transcription has been shown to affect global folding of structural RNAs, such as riboswitches, which in turn modulates the output of gene expression7,20,21,22,23,28. Therefore, we next evaluated how the proximal RNAP modulates the global docking dynamics when paused at G104 (PEC-104), using smFRET between fluorophores attached to P1 and P3 of a hybridized two-strand riboswitch (Fig. 3a and Supplementary Fig. 1), as previously established for the isolated aptamer with stabilized P1.113,15.
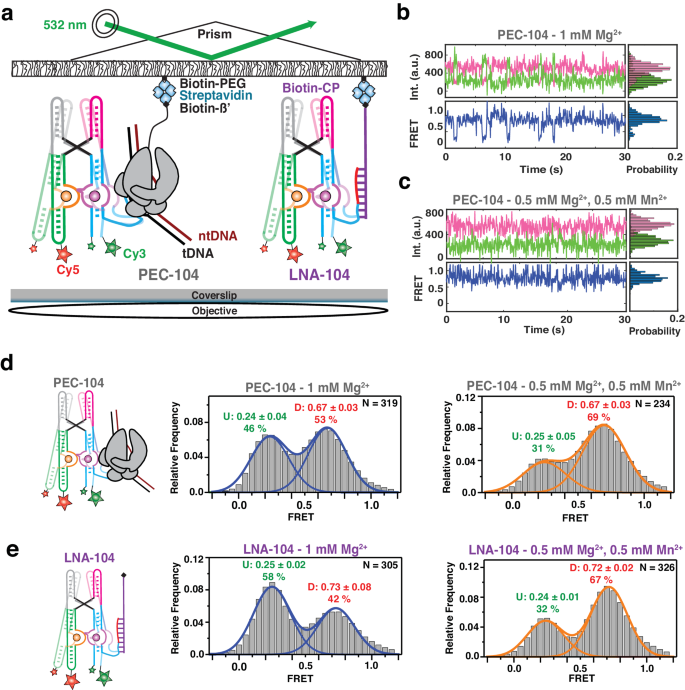
a smFRET experimental setup. Paused Elongation Complex (PEC) immobilization utilizes biotinylated E. coli RNAP. Locked Nucleic Acid (LNA) immobilization utilizes a complementary biotinylated LNA capture probe. Location of donor (Cy3 â green) and acceptor (Cy5 â red) fluorophores are indicated. Representative donor-acceptor (top) and smFRET (bottom) traces for PEC-104 in the absence (b) and presence (c) of 0.5âmM Mn2+. Probability FRET histogram for each trace is indicated on the right. smFRET histograms of PEC-104 (d), LNA-104 (e) in the presence of only Mg2+ (middle panel) and in the presence of both Mg2+ and Mn2+ (right panel). Cartoon of each construct is indicated on the left. The SD of each FRET value is reported. The total number of traces (N) included in each histogram is indicated on top right corner. Source data are provided with this paper at https://doi.org/10.7302/22513.
At near-physiological concentration of Mg2+ (1âmM), the riboswitch in PEC-104 sampled two predominant conformational states in which the low- (~0.2) and high-FRET (~0.7) populations represent the undocked and docked states, respectively (Fig. 3b, d), consistent with previous studies of the isolated aptamer13,15. The FRET histogram showed nearly equal populations in the low- and high-FRET regimes (46% and 53%, respectively), indicating that the riboswitch can occupy the docked state significantly in the absence of Mn2+. Upon addition of 0.5âmM Mn2+, the riboswitch adopted a more stably docked conformation (Fig. 3c) with the high-FRET population increasing to ~70%, indicating that binding of Mn2+ shifts the equilibrium toward the docked state (Fig. 3d and Supplementary Table 4), as previously observed absent the PEC13,15.
To assess the influence of RNAP on the global dynamics of (un)docking, we assembled the same FRET-labeled riboswitch and immobilized it to the microscope slide through a biotinylated locked nucleic acid (LNA) capture probe using the same base pairing employed to reconstitute PEC-104 (LNA-104; Fig. 3a). As illustrated by the presence of the two predominant low- (~0.2) and high-FRET (~0.7) populations, the riboswitch in the absence of RNAP adopted the same two dynamic conformational states as in PEC-104 (Fig. 3e). Notably, in the presence of Mg2+ ions only, the docked population without RNAP was significantly less populated, with 42% for LNA-104 compared to 53% under the same conditions for PEC-104 (Fig. 3d, e), suggesting that the proximal RNAP promotes the formation of the docked state. Similarly, addition of 0.5âmM Mn2+ led to 65% docked state for LNA-104, but a slightly increased 67% for PEC-104 (Fig. 3d, e); whereas in 100âmMâK+ without divalents the riboswitch adopted only 24% docked state for LNA-104, which rose to 35% for PEC-104 (Supplementary Fig. 3 and Supplementary Table 4). These observations demonstrate that RNAP favors docking under a broad set of conditions.
As control, we designed an RNA that went ten bases beyond the G104 pause, and immobilized it through a complementary LNA capture strand (LNA-114) to mimic the structure after full transcription of the riboswitch with RNAP far downstream (Supplementary Fig. 10). In the presence of Mg2+ only, just 25% of the LNA-114 riboswitch were found in the high-FRET (~0.7) docked state, compared to 42% for LNA-104 and 53% for PEC-104 (Supplementary Table 4), suggesting that extension beyond the G104 pause disfavors docking. As expected, with the addition of Mn2+ the fraction of docked state significantly increased to 67 % (Supplementary Fig. 10 and Supplementary Table 4), indicating that the full-length riboswitch remains highly Mn2+ sensitive, even as RNAP has already translocated further downstream.
Taken together, our analysis so far reveals that the presence of RNAP at the G104 pause site promotes the docked riboswitch conformation, particularly in the absence of ligand, due to dynamic sampling of a partially folded P1.1 switch helix.
A semi-docked riboswitch conformation represents the initial nucleation of P1.1
From our previous work it is known that binding of Mn2+ stabilizes a static docked (SD) conformation in the riboswitch in the presence of a complete P1.1 stem13. Further inspection of the smFRET trajectories in our 104 constructs revealed that HMM modeling was best performed using three FRET states (Undockedâ=âU (~0.2), Semi-Dockedâ=âD* (~0.45), Dockedâ=âD (~0.75)). In the presence of Mg2+ only, ~70% of all riboswitch molecules in PEC-104 sampled these three FRET states dynamically over minor stable undocked (SU, ~9%) and SD populations (~20%; Fig. 4a, b). Transitions from the U to the D state (kdock) and from D to U (kundock) occurred at comparable rate constants of ~2.2âsâ1 and ~2.4âsâ1, implying facile reversibility of docking. In the absence of RNAP in LNA-104, the rate constants shifted to ~1.1âsâ1 (kdock) and ~5.5âsâ1 (kundock; Fig. 4e, f), consistent with our previous conclusion that RNAP promotes docking. Likewise, transitions from U or D to D* and from D* to U or D were more prominent in PEC-104 than in LNA-104, suggesting that the semi-docked D* is also favored by the RNAP. A relatively slow kdock,UâD* transition of ~1.8âsâ1 and a faster kundock,D*âU of ~2.9âsâ1 in PEC-104, combined with an even faster kdock,D*âD of ~3.03âsâ1, suggest that the D* state is a frequent, albeit non-obligatory, intermediate between the undocked and undocked states. These findings show that, in the presence of a near-physiological Mg2+ concentration, the riboswitch within the PEC can sample the previously unobserved semi-docked state D*, in addition to the previously reported U and D states.
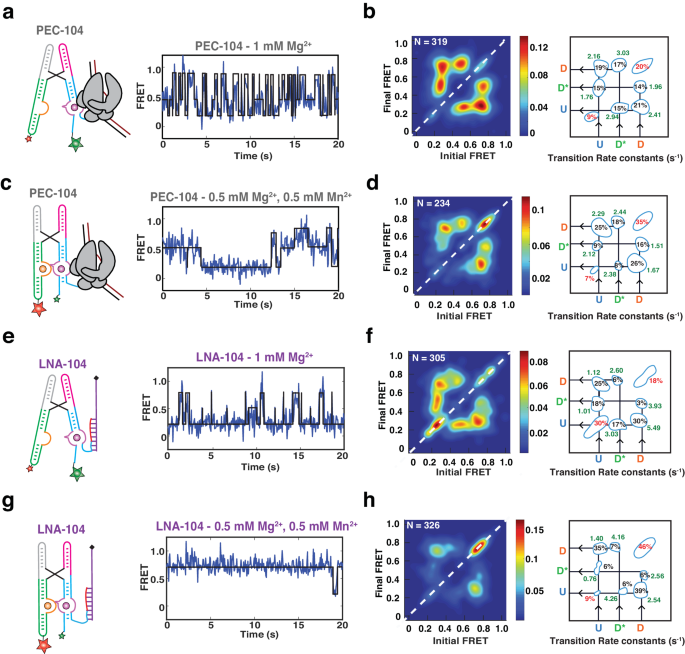
Representative FRET trajectories for PEC-104 in the presence of only Mg2+ (a) and in the presence of both Mg2+ and Mn2+ (c) along with the corresponding Transition Occupancy Density Plots (TODP) and the transition rate constants calculated in the absence (b) and in the presence (d) of Mn2+, See also Supplementary Figs. 5 and 6. eâh Representative FRET trajectories for LNA-104 in the presence of only Mg2+ (e) and in the presence of both Mg2+ and Mn2+ (g) along with the corresponding Transition Occupancy Density Plots (TODP) and the transition rate constants calculated in the absence (f) and in the presence (h) of Mn2+, See also Supplementary Figs. 8 and 9. TODPs represent dynamic traces as âoff-diagonalâ and the static traces as âon-diagonalâ contour, where the color scale shows the prevalence of each population. The riboswitch conformational state corresponding to each FRET value is indicated. Uâ=âUndocked, D*â=âPre-Docked, Dâ=âDocked. Source data are provided with this paper at https://doi.org/10.7302/22513.
As expected from the FRET population histograms, addition of Mn2+ increased the proportion of the on-diagonal SD population in both PEC-104 (from ~20% to ~35%; Fig. 4c, d) and LNA-104 (from ~18% to ~46%). As a result, the dynamic population transitioning between the U, D* and D states decreased to ~58%, supporting that ligand binding shifts the equilibrium toward a more compact, stably docked conformation. Indeed, the transition rate constant from U to D slightly increased from 2.16âsâ1 in the absence to 2.29âsâ1 in the presence of Mn2+, while the transition from D to U became slower (2.41âsâ1 and 1.67âsâ1 in the absence and presence of Mn2+, respectively). Binding of Mn2+ significantly shifted the transition towards higher D* and D populations, with a smaller population now sampling the U state (Fig. 4d). Similarly, the U to D* transitions became faster in the presence of Mn2+ (2.12âsâ1 compared to 1.76âsâ1 with only Mg2+), whereas the transition from D to D* became slower (1.51âsâ1 compared to 1.96âsâ1 with only Mg2+), further supporting that the D* state represents a semi-docked state on route to Mn2+-induced full docking.
In the absence of RNAP (LNA-104), the riboswitch was less dynamic than in PEC-104, preferentially adopting the SU (~30%) and SD (~46%) populations in the absence and presence of Mn2+, respectively (Fig. 4eâh). In addition, kdock was slower and kundock faster than in the absence than the presence of RNAP (Fig. 4f, h), further supporting that proximal RNAP promotes the adoption of the D state. LNA-104 docking was closer to a two-state type transition than PEC-104, with the D* state somewhat less populated. This was particularly true in the presence of Mn2+, in which the D* to D transitions increased from 2.44âsâ1 in the presence to 4.16âsâ1 in the absence of RNAP. This observation suggests that the RNAP can stabilize the semi-docked state, possibly through electrostatic and steric interaction with the riboswitch as observed in the preQ1-sensing riboswitch PEC from B. subtilis21.
As a further control, we analyzed the FRET transition rate constants in the extended LNA-114 construct in the absence and presence of Mn2+ ion (Supplementary Fig. 11). With a now-fully available P1.1 switch helix, the riboswitch behaved similarly to LNA-104, exhibiting a significant proportion of SU (42%) and SD (40%) in both the absence and presence of Mn2+, respectively. In addition, the D* state was observed to be much less accessed than in both LNA-104 and PEC-104, consistent with the notion that the semi-docked conformational state D* may depend on an only partially folded P1.1. Even with three-state HMM fits, the TODP predominantly exhibited two-state transitions between the U and D states (Supplementary Fig. 11). In fact, the overall behavior resembled that of a manganese-sensing riboswitch with a stabilized P1.113, with kundock becoming slower upon ligand binding. Similarly, kundock,D*âU in the presence of Mn2+ was significantly faster for LNA-114 (7.14âsâ1) than LNA-104 (4.26âsâ1), indicating a less stable D* state when the P1.1 is fully stabilized. This observation also explains why D* was not significantly detected in the earlier study featuring the stabilized P1.113.
To further test whether the observed conformational changes are due to stabilization of switching helix P1.1 in PEC-104, we directly surveyed P1.1 folding using smFRET. To measure dynamic distances along the P1.1 helical axis, we employed stepwise transcription to fluorescently label the 5âend of the transcript with Cy3 and incorporate an azido-modified NTP at position 11 (Supplementary Fig. 1), which subsequently was labeled using click-chemistry with DBCO-Cy522,55. To stall the RNAP at position 104 (generating PEC-104), the DNA template was immobilized on streptavidin-coated magnetic beads using 3âdesthio-biotin, which allowed for subsequent bead release via a competing biotinylated-DNA oligonucleotide (Supplementary Fig. 12).
At near-physiological concentration of Mg2+ only (1âmM), helix P1.1 in PEC-104 sampled two predominant conformational states of mid- (~0.40) and high-FRET (~0.65) values and nearly equal populations (58% and 42%, respectively; Supplementary Fig. 13b). Upon addition of 0.5âmM Mn2+, P1.1âs high-FRET state became more prominent (Supplementary Fig. 13c), suggesting that it is associated with the tertiary docked state that is similarly stabilized by Mn2+, consistent with our previous observations on the other FRET vector (Fig. 3). Notably, the majority of traces were static in one or the other FRET state, populating on-diagonal contours in the TODPs (Supplementary Fig. 13d, e). This finding may indicate either very fast transitions beyond the time resolution of our smFRET acquisition, similar to the short-lived (3âms) rare (âexcitedâ) state of the fluoride-sensing riboswitch from B. cereus56, or long-lived steric clashes with the nearby RNAP surface.
Taken together, our single-molecule kinetic analyses reveal a previously unobserved semi-docked, pre-folded riboswitch conformation involving the initial nucleation of P1.1 and stabilization by both RNAP and ligand binding.
Transcription factor NusA recruitment stabilizes P1.1 but is ejected upon ligand binding
Our single-molecule and bulk data suggest a mechanism wherein the initial nucleation of P1.1 extends intrinsic RNAP pausing, allowing time for co-transcriptional folding of the riboswitch into its ligand-sensing conformation for gene regulation. Transcription factor NusA is known to promote transcriptional pausing in the context of hairpin-stabilized Class I pauses through interaction with both the nascent transcript and the RNAP44,57,58. Because folding of P1.1 within the RNAP exit channel could act similarly to a Class I transcriptional pause, we next evaluated RNAP pausing efficiency in the presence of NusA (Fig. 5a). Indeed, in the presence of Mg2+ only, we observed a ~4-fold increase of the G104 half-life upon addition of NusA (210âs in the absence of versus 906âs in the presence of NusA; Fig. 5b, c and Supplementary Table 2). In contrast, in the presence of Mn2+ NusA no longer has any effect on the G104 pause half-life (859âs in the absence versus 820âs in the presence of NusA). These findings suggest that the ligand-bound riboswitch with its stabilized P1.1 helix supersedes NusA-mediated enhancement of G104 pausing efficiency, as also seen for the fluoride-sensing riboswitch23, suggesting redundant action by Mn2+ and NusA.
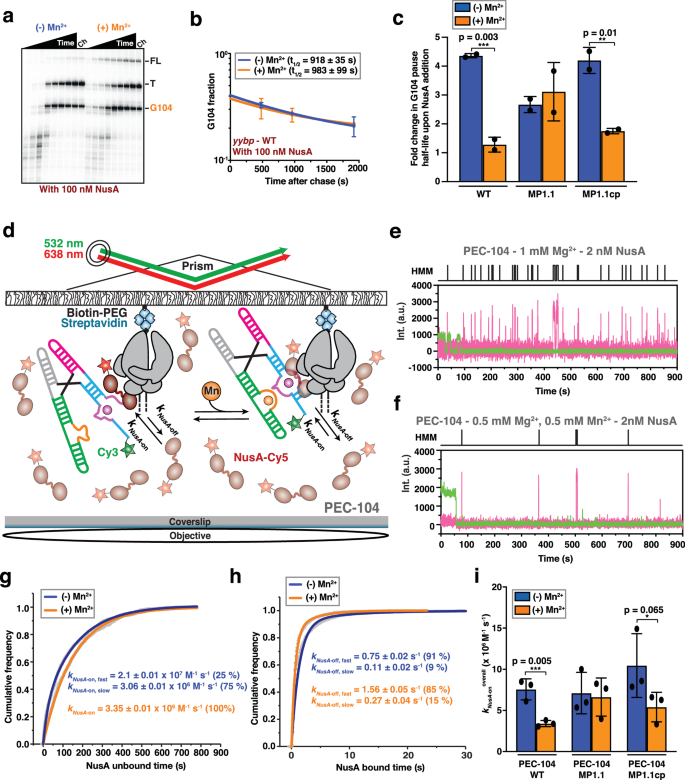
a Representative denaturing gel showing the RNAP pauses during the transcription of the Mn2+ riboswitch in the presence of 100ânM NusA transcription factor. Position of the pause (G104), termination (T) and full-length (FL) products are indicated on the right. Experiments were performed using 25âµM rNTPs in the absence (âMn2+) and presence (+Mn2+) of 0.5âmM Mn2+. The chase lanes (Ch) were taken at the end of the time course after an additional incubation with 500âµM rNTPs for 5âmin. Unprocessed images are provided in the Supplementary Information. b Fraction of complexes at the G104 pause in the presence of NusA transcription factor as a function of the transcription time in the absence (blue) and presence (orange) of 0.5âmM Mn2+. The reported errors are the SD of the mean from nâ=â2 independent replicates. c Fold-change enhancement of G104 pause half-life in the WT, MP1.1 and MP1.1cp riboswitch variants obtained in the presence of 100ânM NusA factor relative to the same condition in the absence of the transcription factor. Experiments were performed using 25âµM rNTPs in the absence (blue) and presence (orange) of 0.5âmM Mn2+. Error bars are the SD of the mean from nâ=â2 independent replicates. The statistical significance of differences was determined using the two-tailed Studentâs t-test (***pâ<â0.01, **pâ<â0.05, *pâ<â0.1). d Single-molecule setup for studying colocalization of NusA factor labeled with a single Cy5 fluorescent dye (NusA-Cy5) with a PEC. Immobilization of the PEC on the microscope slide occurs through the biotin-streptavidin roadblock. Repeated bindings of NusA-Cy5 are monitored through direct excitation of the Cy5 fluorescent dye. Representative single-molecule trajectories showing the binding of NusA-Cy5 (pink) to PEC-104 recorded in the absence (e) or presence (f) of 0.5âmM Mn2+ added co-transcriptionally. Hidden Markov modeling (HMM) is indicated on the top of each trace. A.U. arbitrary unit. Plots displaying the cumulative unbound (g) and bound (h) dwell times of NusA-Cy5 in the absence (blue) and presence (orange) of 0.5âmM Mn2+ in the context of PEC-104. The binding (kon) and dissociation (koff) rate constants of NusA are indicated. The reported errors are the error of the fit. Total number of molecules analyzed for each condition is as follow: (â) Mn2+â=â302; (+) Mn2+â=â294. i Overall binding rate constants (kon) of NusA-Cy5 in the context of PEC-104 WT, MP1.1 and MP1.1cp constructs determined in the absence (blue) and presence (orange) of 0.5âmM Mn2+. Error bars are the SD of the mean from nâ=â2 independent replicates. The statistical significance of differences was determined using the two-tailed Studentâs t-test (***pâ<â0.01, **pâ<â0.05, *pâ<â0.1). See also Supplementary Fig. 13. Total number of molecules analyzed for each condition is as follow: PEC-104 WT (â) Mn2+â=â302; PEC-104 WT (+) Mn2+â=â294; PEC-104 MP1.1 (â) Mn2+â=â232; PEC-104 MP1.1 (+) Mn2+â=â303; PEC-104 MP1.1cp (â) Mn2+â=â276; PEC-104 MP1.1cp (+) Mn2+â=â301. Source data are provided with this paper at https://doi.org/10.7302/22513.
Because we identified the P1.1 switch helix as the key modulator of the G104 pause in the absence of the transcription factor (Fig. 1d), we next tested the hypothesis that it might also negotiate NusAâs ability to affect transcriptional pausing. Indeed, while the MP1.1 mutant completely abolished the effect of Mn2+ on pausing efficiency (Fig. 1d), the mutant allowed NusA to retain its ability to extend pausing independent of Mn2+ (Fig. 5c). Complementarily, when the compensatory mutant MP1.1cp was used, the wild-type behavior was restored, i.e., NusAâs large effect on pausing in Mg2+ only was lost upon addition of Mn2+ (~4-fold and only ~1.5-fold enhancement of G104 pause half-life by NusA without and with ligand, respectively; Fig. 5c and Supplementary Table 2). These data further support that Mn2+ and NusA have a redundant function in P1.1 formation. Interestingly, addition of NusA during in vitro transcription of the riboswitch moderately decreased the concentration of Mn2+ needed to modulate transcription termination and readthrough (Supplementary Fig. 14a, b), suggesting that NusA may not only alter the transcription rate and thus time window for ligand sensing, but also modestly sensitize the riboswitch to its ligand17,23,51. In further support, an in vitro termination assay performed in either the absence or presence of NusA revealed a small, but significant decrease in transcription readthrough in the presence of both ligand and transcription factor, from 60% in the absence to 43% in the presence of NusA (Supplementary Fig. 14c).
To understand the mechanism by which NusA and Mn2+ alternatively affect G104 pause behavior, we directly monitored the dynamics of NusAâs interaction with PEC-104 through an established single-molecule colocalization assay (Fig. 5dâf)23. In the presence of Mg2+ only, NusA bound with two rate constants of 2.10â±â0.01 à 107 Mâ1 sâ1 (kNusA-on,fast; fraction 25%) and 3.06â±â0.01 à 106 Mâ1 sâ1 (kNusA-on,slow; fraction 75%), as well as two dissociation rate constants of 0.75â±â0.02âsâ1 (kNusA-off,fast; fraction 91%) and 0.11â±â0.02âsâ1 (kNusA-off,slow; fraction 9%; Fig. 5g, h and Supplementary Table 5). Similar to the SiM-KARTS probe, these biphasic kinetics suggest that P1.1 exists in (at least) two conformational states that NusA senses. Interestingly, the binding kinetics resemble those determined for NusA binding to a canonical Class I pause PEC (hisPEC)23, further supporting that the G104 pause is stabilized through the formation of the P1.1 switch helix within the RNAP exit channel.
In the presence of Mn2+, the cumulative unbound dwell times could only be fitted with a single-exponential kNusA-on of 3.35â±â0.01 à 106 Mâ1 sâ1 (Fig. 5g), similar to the slower binding rate constant in Mg2+ only, indicating that Mn2+ binding leads to a more homogenously folded P1.1 that inhibits the fast recruitment of NusA to PEC-104. The still two dissociation rate constants increased by ~2-fold upon addition of ligand, suggesting a competition between Mn2+ and NusA binding. Overall, these observations demonstrate that ligand binding affects the early step of NusA-mediated enhancement of pausing by preventing initial recruitment of NusA to PEC-104. We were able to link these observations again to P1.1 switch helix formation, since the effect of Mn2+ on NusA binding was lost for the MP1.1 mutant and largely restored for the compensatory MP1.1cp mutant (Fig. 5i, Supplementary Fig. 15 and Supplementary Table 5).
To ask whether the observed effect of ligand binding on NusA recruitment occurs during co-transcriptional folding, we next surveyed NusA binding during real-time transcription of the riboswitch23,59 (Supplementary Fig. 16). In Mg2+ only, and absence of Mn2+, NusA remains bound to the active transcription complex for ~1.8âs, similarly to our results on the stalled PEC-104 under the same conditions (Supplementary Table 5). In contrast, when Mn2+ is added co-transcriptionally, a slight, but significant decrease of the bound time is observed (~1.2âs; Supplementary Fig. 16c), indicating that co-transcriptional folding of the riboswitch in the presence of ligand alters NusA occupancy to the transcription complex in situ.
Collectively, our single-molecule and biochemical assays demonstrate that NusA-mediated enhancement of transcriptional pausing is abolished by Mn2+-induced nucleation of P1.1, which in turn suppresses NusA recruitment to the PEC. That is, while functionally redundant in extending transcriptional pausing by favoring a stable P1.1, Mn2+ and NusA do so in mutually exclusive fashion.
NusA stabilizes the transient semi-docked riboswitch conformation
Our single-molecule NusA binding assay unveiled that ligand binding to the riboswitch prevents fast NusA recruitment to the PEC through P1.1 stabilization. While NusA binds transiently, as one of the most abundant transcription factors in the bacterial cell (at micromolar concentrations)60 it is expected to be almost continuously bound to an active elongation complex (EC) in vivo61 so that it is unlikely that PEC-104 exists in the absence of NusA. To understand the dynamics of the riboswitch in a context closer to in vivo conditions, we next evaluated the global docking dynamics of the NusA-bound PEC-104 using smFRET in the absence and presence of Mn2+ (Fig. 6a).
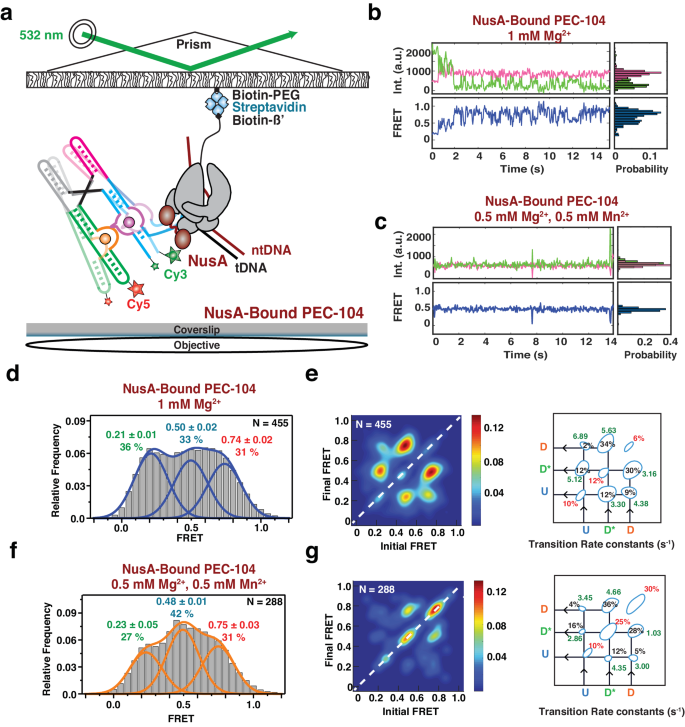
a smFRET experimental setup. Paused Elongation Complex (PEC) immobilization utilizes biotinylated E. coli RNAP. Location of donor (Cy3 â green) and acceptor (Cy5 â red) fluorophores are indicated. Representative donor-acceptor (top) and smFRET (bottom) traces for NusA-Bound PEC-104 in the absence (b) and presence (c) of 0.5âmM Mn2+. Probability FRET histogram for each trace is indicated on the right. SmFRET histograms of NusA-bound PEC-104 in the presence of only Mg2+ (d) along with the corresponding Transition Occupancy Density Plots (TODP) and the determined transition rate constants (e). SmFRET histograms of NusA-bound PEC-104 in the presence of both Mg2+ and Mn2+ (f) along with the corresponding Transition Occupancy Density Plots (TODP) and the determined transition rate constants (g). The SD of each FRET value is reported. The total number of traces (N) included in each histogram is indicated on top right corner. See also Supplementary Figs. 16 and 17. TODPs represent dynamic traces as âoff-diagonalâ and the static traces as âon-diagonalâ contour, where the color scale shows the prevalence of each population. The riboswitch conformational state corresponding to each FRET value is indicated. Uâ=âUndocked, D*â=âPre-Docked, Dâ=âDocked. Source data are provided with this paper at https://doi.org/10.7302/22513.
Similar to the traces collected for PEC-104 in the absence of Mn2+, i.e., in the absence of any divalent ion or with Mg2+ only, the NusA-saturated PEC-104 shows a high degree of dynamics between the three FRET states U, D* and D (Fig. 6b and Supplementary Fig. 17). In contrast, upon addition of ligand, the smFRET traces become more static, preferentially adopting the D* or D states (Fig. 6c), suggesting that both NusA and ligand stabilize the semi-docked and docked conformation of the riboswitch. Accordingly, in both the absence and presence of Mn2+ with NusA bound the cumulative FRET histograms could be best fitted with three independent Gaussian peaks, corresponding to the three conformational states (U, D* and D; Fig. 6d, f). Moreover, upon Mn2+ addition, the equilibrium shifted from the U toward the D* state, rather than adopting the D state, further supporting that NusA particularly favors this intermediate semi-docked state.
Kinetic analysis of the transitions between the different conformational states further confirmed that the riboswitch was mostly dynamic in the absence of ligand. However, while the proportion of static molecules remained the same (29% in the absence versus 28% in the presence of NusA) with only Mg2+, the transitions from U to either the D* or D states became much faster in the NusA-bound PEC compared to PEC-104 in the absence of the transcription factor (compare Fig. 6e and Fig. 4b and Supplementary Figs. 5 and 19). Indeed, both kdock,UâD and kdock,UâD* significantly increased in the presence of saturating transcription factor, suggesting that NusA promotes fast dynamics into both states (Fig. 6e). Further, the transitions between the D* and D states were observed to be the most probable (64%) with a faster regime toward the D state (5.63âsâ1; fraction 34% in the presence of NusA versus 3.03âsâ1; fraction 17% in the absence of NusA).
Upon addition of Mn2+, the transition kinetics between the conformational states revealed that a 25% static population was found in SD* and 30% in SD (Fig. 6g), suggesting that NusA assists Mn2+-induced stable docking. Most of the dynamic transitions (~64%) were observed between the D* and D states, with faster transitions toward the D state compared to PEC-104 without NusA-bound (4.66âsâ1; fraction 36% in the presence of NusA versus 2.44âsâ1; fraction 18% in the absence of NusA; Fig. 6g and Supplementary Fig. 20). Conversely, the transitions from the D to the D* state were significantly slower in the presence of both Mn2+ and NusA (1.03âsâ1; fraction 28% in the presence of NusA versus 1.51âsâ1; fraction 16% in the absence of the transcription factor), supporting that NusA promotes full docking of the riboswitch through stabilization of the P1.1 switch helix. In further support, SiM-KARTS probing of P1.1 performed on NusA-bound PEC-104 revealed that this region is less accessible in the presence of the transcription factor (Supplementary Fig. 21 and Supplementary Table 3), confirming its impact factor on the local stabilization of P1.1.
NusA preferentially recognizes the single-stranded P1.1 in the ligand-free conformation
Our single-molecule binding assays and structural probing so far unveiled a previously unappreciated effect of NusA transcription factor on nascent RNA structural dynamics and vice versa (Figs. 5 and 6). While the P1.1 switching helix is central for the overall gene regulation mediated by ligand binding, from transcriptional pausing to transcription termination, it is still unclear which conformational state is preferred for the initial recruitment of NusA. To probe the mechanistic details of NusA-mediated regulation of gene expression as a function of RNA structure, we next evaluated the kinetics of NusA binding to an âopenâ P1.1 helix using a 3-color microscopy experiment. Here, the open (single-stranded) state of P1.1 was surveyed through Cy3-labeled SiM-KARTS probe binding (green), while simultaneously observing the binding of Cy5-labeled NusA (pink) to PEC-104 (blue, Fig. 7a, b).
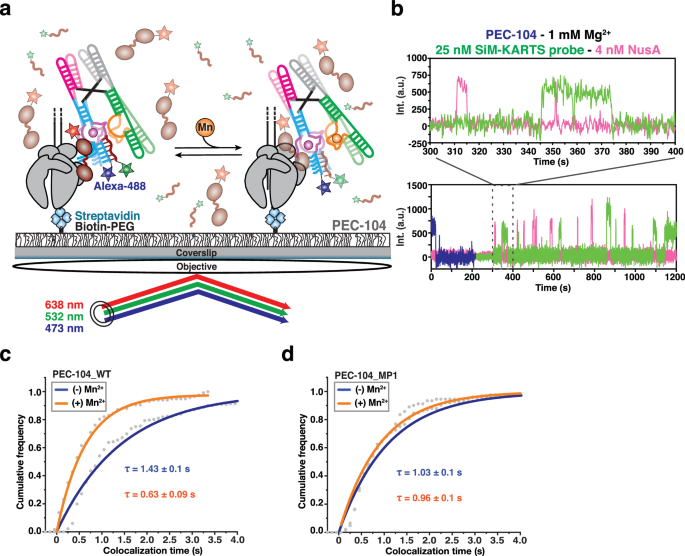
a Single-molecule setup for studying colocalization of NusA factor labeled with a single Cy5 fluorescent dye (NusA-Cy5) and SiM-KARTS probe labeled with a single Cy3 fluorescent dye with a PEC. Immobilization of the PEC on the microscope slide occurs through the biotin-streptavidin roadblock. Repeated bindings of NusA and SiM-KARTS probe are monitored through direct excitation of the Cy3 and Cy5 fluorescent dyes. b Representative single-molecule trajectories showing the binding of NusA-Cy5 (pink) and SiM-KARTS probe (green) to single PEC-104 (blue). A.U., arbitrary unit. Plots displaying the cumulative colocalization time NusA-Cy5 and the SiM-KARTS probe in the absence (blue) and presence (orange) of 0.5âmM Mn2+ in the context of PEC-104_WT (c) and PEC-104_MP1 (d). The colocalization dwell times are indicated. The reported errors are the errors calculated from bootstrapping of all dwell times collected. Total number of colocalization events analyzed for each condition is as follow: PEC-104_WT (â) Mn2+â=â232; PEC-104_WT (+) Mn2+â=â117; PEC-104_MP1 (â) Mn2+â=â124; PEC-104_MP1 (+) Mn2+â=â91. Source data are provided with this paper at https://doi.org/10.7302/22513.
In the presence of only Mg2+, NusA colocalized with the undocked P1.1 helix for ~1.4âs, a value very similar to the overall bound time of NusA to PEC-104 under the same conditions (Supplementary Table 5), supporting the notion that NusA captures the open P1.1 during its recruitment. Strikingly, upon addition of Mn2+, this colocalization dwell time significantly decreased by ~2.5 fold (Fig. 7c), suggesting that folding of the P1.1 helix in the presence of ligand and NusA occupancy are mutually exclusive. Control experiments surveying colocalization dwell times in the MP1 mutant of PEC-104 further corroborated that the effects on NusA association kinetics observed upon ligand binding require the ability to form the top base pairs of P1.1 (Fig. 7d).
Taken together, we discovered a previously unappreciated effect of NusA transcription factor on RNA structural dynamics, wherein binding of NusA to PEC-104 stabilizes an intermediate, semi-docked state of the riboswitch to assist downstream folding in the ligand-bound conformation. All of these effects involve the P1.1 switch helix as a focal point on which Mn2+, NusA and RNAP all act; in return, P1.1 integrates these disparate signals as it modulates gene expression.