Sampling site
Subsurface seawater (1 m depth) was collected at 52 time points between 2nd of March and 26th of May 2020 at the station Helgoland Roads near Helgoland in the southern North Sea. Since 1962 bucket water samples have been taken as part of a long-term monitoring program Helgoland Roads (54°11âN 7°54âE; DEIMS.iD: https://deims.org/1e96ef9b-0915-4661-849f-b3a72f5aa9b1)42.
Sequence analysis
Sequence analysis was carried out on the basis of the existing annotation20 with additional reannotation as describes previously21. Sequences were aligned using ClustalO (v2.1)43 in Unipro UGENE (v47)44. Trees were visualized using iTOL (v6.8.1)45.
Chlorophyll a measurements and cells counts of total bacteria and dominant bacterial clades
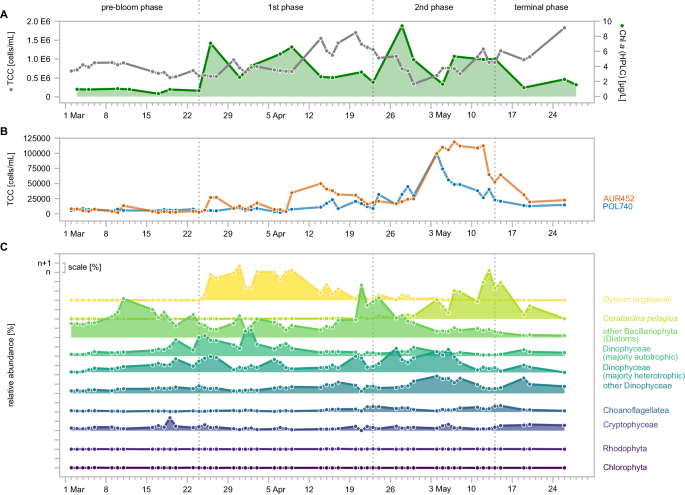
Sample filtration was carried out under dim light to avoid the pigment loss during the filtration procedure. For 2016 and 2018 samples, pigment extraction and analysis was carried out using a combined protocol from Zapata et al.46 and Garrido et al.4,47. For 2020 samples, we followed the extraction and analysis method as described previously48. Subsequently, pigments were separated via high-performance liquid chromatography (HPLC) (Waters 2695 Separation Module), and detected with a Waters 996 Photodiode Array Detector (Waters, Milford, MA, USA). Total bacterial cell counts (TCC) and cell numbers of the dominant clades Aurantivirga (CARD-FISH probe AUR452) and Polaribacter (POL740) of the 2020 spring phytoplankton bloom were described and published previously17.
18S rRNA gene sequencing
Sampled water was sequentially filtered onto polycarbonate membrane filters with different pore sizes (10âμm, 3âμm and 0.2âμm). For 18S rRNA gene amplicon sequencing, the two larger size fractions, >10âμm and 3â10âμm were analyzed. DNA was extracted from the filters using the DNeasy PowerSoil kit for DNA (Qiagen GmbH, Hilden, Germany). Mechanical lysis was achieved by bead beating in a FastPrep 24 5G (MP Biomedicals LLC, Irvine, CA, USA). The V7 region of the 18S rRNA gene was amplified using the primers [F-1183mod: 5â-AATTTGACTCAACRCGGG-3â, R-1443mod: 5â-GRGCATCACAGACCTG-3â]49 coupled to custom adapter-barcode constructs. PCR amplification and Illumina MiSeq library preparation (Illumina Inc., San Diego, CA, USA) and sequencing (V3 chemistry) was carried out by LGC Genomics in Berlin. Sequences have been submitted to the European Nucleotide Archive under the accession number PRJEB51816. Amplicon Sequence Variants (ASVs) were obtained using DADA2 (v1.26)50 and taxonomically classified as described previously51. ASVs classified as Metazoa were removed before downstream analyses to reduce the effect of in particular crustacean zooplankton on community composition. For analysis, 10âμm and 3â10âμm counts were combined, set to 1 and relative abundances calculated. Classification of Dinophyceae into majorly heterotroph and majorly autotroph taxa was done for dominant groups according to literature13,52,53,54,55.
Metatranscriptomics
Metagenome and metatranscriptome sequencing were performed as described previously17. Briefly, thirty metagenomes were sequenced using PacBio Sequel II (Pacific Biosciences, Menlo Park, CA, USA) with one SMRT cell/sample while corresponding metatranscriptomes were obtained using Illumina HiSeq 3000 (~100 million reads/sample). The metagenomes were then processed to reconstruct metagenome-assembled genomes (MAGs), and mRNA reads were mapped to these genomes to identify highly expressed MAGs. Mapping and annotation were carried out using SqueezeMeta v1.3.156. To predict open reading frames (ORFs), FragGeneScan v1.31 with the parameters âw1â and âsanger_5â as described by Rho et al.57 was used. The predicted ORFs were searched against various databases including GenBank r23958, eggNOG v5.059, KEGG r58.060, and CAZy (as of 11/12/2023)61 using Diamond v0.9.24.12562. HMM homology searches against the Pfam 33.0 database63 were conducted using HMMER364. The combined annotations were utilized for the manual prediction of polysaccharide utilization loci (PULs) and carbohydrate-active enzyme (CAZyme) clusters. Bowtie265 was employed to map mRNA reads to the MAGs, and transcripts per million (TPM) values were calculated for all MAGs in a given sample using the formula: (sum of reads successfully mapping to a MAG in the sample Ã10^6)/(sum of contig lengths of the MAGâÃâsum of reads in the sample).
Metaproteomics
Sample preparation
The metaproteomics analysis of the 0.2âµm fraction from the spring phytoplankton bloom in 2016 has been described in detail in21 and the free-living bacteria of the blooms from 2018 and 2020 were prepared as described previously66. Briefly, proteins were extracted from one-eighth of a filter (Millipore Express PLUS Membrane, polyethersulfone, hydrophilic, 0.2âµm pore size, diameter 142âmm) by cutting the filter into small pieces before transfer to 15âmL low binding tubes containing 1âmL resuspension buffer 1 (50âmM Tris-HCl (pH 7.5), 0.1âmg mLâ1 chloramphenicol, 1 mM phenylmethylsulfonyl fluoride (PMSF)) and 1,5âmL resuspension buffer 2 (20âmM Tris-HCl pH 7.5, 2% SDS (w/v)). After heating (10âmin at 60â°C at 1000 rpm in a thermo-mixer), 5 mL DNAse buffer (20âmM Tris-HCl pH 7.5, 0.1âmg mLâ1 MgCl2, 1 mM PMSF, 1âμg mLâ1 DNAse I) was added, and cells lysis was carried out by ultra-sonication (amplitude 51â60%; cycle 0.5; 3à 2 min) on ice before incubation for 10âmin at 37â°C at 1000 rpm. After centrifugation (10 min at 4â°C at 10,000âÃâg), the supernatant was collected and the pelleted filter pieces were stirred and centrifuged again for 1 min at 4â°C at 5000âÃâg. Pre-cooled trichloroacetic acid (20% TCA (v/v)) was added for protein precipitation to the supernatant and after inverting the tube approximately 10x, the precipitate was pelleted via centrifugation (45âmin, 4â°C, 12,000âÃâg) and the protein pellet was washed 3à in pre-cooled (â20â°C) acetone (10 min, 4â°C, 12,000âÃâg) before drying at room temperature. The proteins were resuspended in 2à SDS sample loading buffer (4% SDS (w/v), 20% glycerol (w/v), 100âmM Tris-HCl pH 6.8, bromphenol blue (tip of a spatula, to add color), 3.6% 2 mercaptoethanol (v/v) (freshly added before use)), incubated for 5âmin at 95â°C before vortexing and separated via SDS-PAGE (Criterion TG 4â20% Precast Midi Gel, BIO-RAD Laboratories, Inc., USA). The proteins were fixated, stained with Coomassie, and each gel lane was cut into 20 pieces67. Gel pieces were destained 3à for 10 min with 1 mL of gel washing buffer (200âmM ammonium bicarbonate in 30% acetonitrile (v/v)) at 37â°C under vigorous shaking, dehydrated in 1âmL 100% acetonitrile (v/v) for 20âmin and the supernatant was removed before drying the gel pieces in a vacuum centrifuge at 30â°C. Proteins were in-gel reduced with 100âµL 10âmM dithiothreitol in 25âmM ammonium bicarbonate buffer (1âh at 56â°C) and alkylated with 100âµL 55âmM iodoacetamide in 25âmM ammonium bicarbonate buffer (without light for 45âmin at room temperature) before the supernatant was removed. The gel pieces were washed with 1âmL 25âmM ammonium bicarbonate buffer (10âmin, 1000 rpm at room temperature), dehydrated with 500âµL (2018 bloom) or 800âµL (2020 bloom) 100% acetonitrile for 10âmin. The supernatant was removed before gel pieces were dried in a vacuum centrifuge (20 min) and finally covered with 120âµL trypsin solution (2 µg/mL Trypsin (Promega). After incubation for 20âmin at room temperature, excess trypsin solution was removed and incubated in a thermo-mixer 15âh at 37â°C without shaking. Peptides were eluted with 120 µL solvent A (water MS grade in 0,1% acetic acid (v/v)) by sonication for 15 min before protein containing supernatant was transferred into a new tube. Peptide elution was repeated with 120 µL 30% acetonitrile (v/v) by sonication for 15âmin. The eluates were pooled, and eluate volume was reduced in a vacuum centrifuge to a maximum of 15 to 20âµL. The peptides were desalted via ZipTips µC18 (Merck Millipore, P10 tip size) according to the manufacturerâs protocol. The eluted samples were dried in a vacuum centrifuge and resuspended in 10âµL 0.5à Biognosys iRT standard kit in solvent A.
LC-MS/MS measurement and data analysis
Information about the LC MS/MS measurement and data analysis of the 0.2 µm fraction from the spring phytoplankton bloom in 2016 has been described in detail in ref. 21 and are described here for the free-living bacteria of the blooms from 2018 and 2020. An Easy-nLC1000 (Thermo Fisher Scientific, Waltham, MA, USA) was coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific) and peptides were loaded onto in-house packed capillary columns (20âcm length,75âµm inner diameter) filled with Dr. Maisch ReproSil Pur 120 C18-AQ 1.9âµm (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) and separated using a 131 min nonlinear binary gradient from 1% to 99% solvent B (99.9% acetonitrile(v/v), 0.1% acetic acid (v/v)) in solvent A (0.1% acetic acid (v/v)) at a constant flow rate of 300ânL minâ1. The MS1 scan was recorded with a mass window of 300â1650âm/z and a resolution of 140,000 at 200âm/z. The 15 most intense precursor ions were selected for HCD fragmentation (ions with an unassigned charge or a charge of 1, 7, 8, >8 were excluded) with a normalized collision energy of NCE 27. The resulting MS/MS spectra were recorded with a resolution of 17,500 at 200âm/z. Dynamic exclusion and lock mass correction were enabled.
All MS/MS spectra were analyzed using Mascot (version 2.7.0.1; Matrix Science, London, UK) and a bloom-specific metagenome-derived database containing all protein sequences from the 18 metagenomes obtained during the spring bloom in 2018 or 15 metagenomes obtained during the spring bloom 2020 assuming the digestion enzyme trypsin. Redundant proteins were removed using cd-hit68 with a clustering threshold of 97% identity. The non-redundant database was added by a set of common laboratory contaminants and reverse entries, amounting to 81,874,922 (bloom 2018) or 4,221,978 (bloom 2020) sequences in the final database.
For database search with Mascot69, the following parameters were used: fragment ion mass tolerance and parent ion tolerance of 10 ppm, none missed cleavages, variable modification on methionine (oxidation), and fixed modification on cysteine (carbamidomethylation). Scaffold (version 4.11.1 (bloom 2018) or version 5.0.1 (bloom 2020); Proteome Software Inc., Portland, OR) was used to merge the search results and to validate MS/MS-based peptide and protein identifications70. During data analysis in Scaffold, an additional X! Tandem search was performed for validation (version 2017.2.1.4; The GPM, thegpm.org; version X!Tandem Alanine)71 with default settings (fragment ion mass tolerance and parent ion tolerance of 10 ppm, carbamidomethyl on cysteine as fixed modification, Glu->pyro-Glu of the N-terminus, ammonia-loss of the N-terminus, Gln->pyro-Glu of the N-terminus and oxidation on methionine for 2018 and 2020 bloom, and additional carbamidomethyl of cysteine as variable modifications for 2018 bloom). Peptide identifications were accepted if they could be established at greater than 95% probability. Peptide probabilities from Mascot were assigned by the Peptide Prophet algorithm (bloom 2018)72 or the Scaffold Local FDR algorithm (bloom 2020). Peptide Probabilities from X! Tandem were assigned by the Peptide Prophet algorithm72 with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 99% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm73. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
For (semi-)quantitative analysis of 201621, 2018 and 2020 metaproteomic datasets, percent normalized weighted spectra (%NWS) were calculated by dividing Scaffoldâs âQuantitative Valueâ for normalized, weighted (i.e. protein size-adjusted) spectra for each protein group, by the sum of all quantitative values for the sample. Average values were calculated from three biological replicates, including â0â for proteins that were not identified within a replicate. To make bacteria-specific %NWS readily comparable across all samples, all bacterial spectra were normalized to 100% (%BacNWS) using taxonomic assignment for protein groups provided by GhostKOALA v2.074 (genus_prokaryotesâ+âfamily_eukaryotesâ+âviruses database).
Comparative genomics
MAGs from 2010-2012, 201621 (European Nucleotide Archive project accession PRJEB28156), 2018 (PRJEB38290) and 202017 (PRJEB52999) were dereplicated using dRep75 v3.2.0 with minimum completeness of 70% and contamination lower than 5% at 0.95 ANI (average nucleotide identity). Protein sequences for representative MAGs were predicted with Prokka76 v1.14.6. PULs, CAZymes, SusC-like and SusD-like proteins were predicted as described previously21 using hmmscan v3.3.2 against dbCAN-HMMdb-V12 and diamond62 v2.1.1.155 against CAZyDB.07262023 provided by dbCAN77. PULs were predicted with a sliding window of seven genes, i.e. the CAZymes and susC/D genes can only be seven genes apart for them to be considered part of the same PUL. GH13-encoding PULs were classified as âα-glucan-targetingâ, whereas PULs carrying combinations of GH3, GH16, GH17, GH30 and/or GH5 enzymes were annotated as âβ-glucan-targetingâ. These substrate predictions were curated manually according to further PUL encoded CAZymes.
For identification of enzymes involved in α-glucan synthesis, K numbers were assigned to each sequence by GhostKOALA v2.074 (genus_prokaryotesâ+âfamily_eukaryotesâ+âviruses) and KofamScan78 (ver. 01/04/2023, KEGG release 106) with an E-valueââ¤â0.01. Proteins with hits for K00963, K00975, K00693, K00750, K16150, K16153, K13679, K20812, K00703, K16148, K16147, K00700 and K16149 were kept as part of the α-glucan synthesis pathway.
Strain and cultivation conditions
We used the North Sea flavobacterial strains Polaribacter sp. Hel_I_88 (isolated from seawater off Helgoland island) and Muricauda sp. MAR_2010_75 (isolated from seawater at Sylt island), as model organisms79. For pre-cultures and polysaccharide extractions Polaribacter sp. Hel_I_88 and Muricauda sp. MAR_2010_75 were grown over night (20â°C, 200 rpm) in Marine Broth (MB 2216, Difco). Polaribacter sp. and Muricauda sp. were tested for growth on specific carbon sources in MPM medium80 containing 0.1% (w/v) of a single poly- or monosaccharide (Glycogen from Oyster: Merck; Pullulan: Thermo Fisher Scientific; Glucose: Roth) or 0.2% (w/v) bacterial polysaccharide extract. Growth was assessed via measurement of optical density at 600ânm. Cultures for proteome analysis were carried out in 25âmL MPM medium using biological triplicates.
Proteomics of pure cultures
Cultures of Polaribacter sp. Hel_I_88 for time series sampling were grown in 100âmL batches. Cultures of Polaribacter sp. Hel_I_88 and Muricauda sp. MAR_2010_75 for extract characterization were grown in 25 mL batches. For time series sampling, 25 mL samples were sequentially filtered through 3 and 0.2âμm polycarbonate filters (à 47âmm, Merck) using a vacuum pump (PC 3002 VARIO, VACCUBRAND) after 16, 24 and 48âh. Filters were stored at â80â°C until further use. Protein was extracted from ¼ of each 0.2âμm filter and prepared for mass spectrometry as described for metaproteomics but using 10% 1D-SDS polyacrylamide gels.
For extract characterization, cultures were grown on either polysaccharide extract, glycogen and alginate (Sigma-Aldrich) (Polaribacter sp.) or xylan from beechwood (Sigma-Aldrich) (Muricauda sp.). After 72âh, 25âmL cultures were harvested via centrifugation at 4000âÃâg and stored at â80â°C until further use. Protein was extracted by resuspending the pellet in 2âmL 50âmM TEAB buffer containing 4% (w/v) SDS. Samples were incubated at 95â°C and 600ârpm for 5âmin, cooled on ice and sonicated (HD/UV 2070, Bandelin, Berlin, Germany) for 5âmin. Debris was removed by centrifugation (14,000âÃâg, 10âmin) and protein concentration was determined using the Pierce BCA Protein Assay Kit (ThermoFischer Scientific). Per sample, 25âμg protein were used. Proteins were separated on a 10% 1D-SDS polyacrylamide gel at 120âV for 90âmin.
Samples were measured using an easy nLCII HPLC system applying a 100âmin gradient coupled to an LTQ Orbitrap Velos mass spectrometer (Resolution 30,000, Scan range 300-1 700) (Thermo Fisher Scientific Inc., Waltham, MA, USA)81. Using MaxQuant82, spectra were matched using a target-decoy protein sequence database with sequences and reverse sequences of Polaribacter sp. Hel_I_88 (NCBI ASM68793v1) or Muricauda sp. MAR_2010_75 (NCBI ASM74518v1) and common laboratory contaminants. A protein and peptide level FDR of 0.01 (1%) with at least two identified peptides per protein was applied. Only proteins that were detected in at least two replicates were classed as identified. Relative iBAQ (intensity based absolute quantification) values were manually calculated from automatically calculated iBAQs. Data and Results are available through the ProteomeXchange Consortium via the PRIDE partner repository (http://proteomecentral.proteomexchange.org)83 with the identifier PXD043390. Statistical analysis for differential expression was performed in Perseus (2.0.1.1)84 using one-way ANOVA followed by post-hoc Tukeyâs HSD test.
Cloning, protein expression and purification
Genes coding for the proteins GH13A (P161_RS0117435), GH13B (P161_RS0117440) and GH13C (P161_RS0117455) of Polaribacter sp. Hel_I_88 (NCBI accession NZ_JHZZ01000001) were codon optimized and synthesized without their signal peptide by de novo gene synthesis (BioCat GmbH, Heidelberg, Germany). They were cloned into pET22b+ in Escherichia coli BL21 (DE3) for protein production. The susD gene (P162_RS13765) of Flavimarina sp. Hel_I_48 (NCBI accession: NZ_JPOL00000000) was amplified from genomic DNA via PCR (primers fwd: GTGTCTCGAGTTAATAACCTGGGTTTTGAGTCAGGTTT; rev: GAGAGGATCCTGAGAAT GATCTTGACGTAACCTTAGAG) and cloned into pET22b+ via restriction/ligation. Proteins were produced in 200âmL LB cultures (30âμg mLâ1 ampicillin) by induction with IPTG and incubation over night at 20â°C. Cells were harvested by centrifugation (5000âÃâg, 20âmin), lysed using BugBuster Protein Extraction Reagent (Merck) (GH13s) or sonication on ice (3âÃâ2âmin, 50% cycle, SusD) and centrifuged (9500âÃâg, 20âmin) to remove debris. Proteins were purified by loading the lysate onto a prepacked 5âmL IMAC column (HisTrap HP 5âmL, Cytiva) equilibrated with IMAC Buffer A (100âmM NaCl, 20âmM Imidazole, 20âmM Tris-HCl, pH 8) using an ÃKTA Pure 25 L (Cytiva). Proteins were eluted with a step gradient of IMAC Buffer B (100âmM NaCl, 500âmM Imidazole, 20âmM Tris-HCL, pH 8). Flavimarina sp. SusD was further purified using size-exclusion chromatography (Superdex 200 Increase 10/300 GL, Cytiva) using SEC-Buffer (100 mM NaCl, 20 mM Tris-HCL, pH 7.4). The GH13 A, B & C proteins were desalted using a prepacked sepharose-based desalting column (HiPrep Desalting 26/10, Cytiva) with PBS Buffer (pH 7.4). All proteins were concentrated via spin columns (Pierce Protein Concentrator PES, 30K MWCO, 2â6âmL, Thermo Fischer).
Enzyme characterization
Activity profiles for all enzymes were generated by 3,5-dinitrosalicylic acid (DNS) reducing-end assay85 as well as fluorophore-assisted carbohydrate electrophoresis (FACE)8. 25 μg purified protein were incubated with 0.5% (w/v) poly-/oligosaccharide (dextran 70, maltose, maltotriose, isomaltotriose, and maltopentose from Roth; laminarin from Laminaria digitata, α-/β-cyclodextrin from Sigma; panose and isopanose from Megazyme) for 24âh. Samples were heat-inactivated at 80â°C for 10âmin and centrifuged (13,000âÃâg, 10âmin) to remove precipitated protein.
For the reducing-end assay, samples were incubated with DNS-reagent solution (30% (w/v) Potassium sodium tartrate tetrahydrate, 10âmg mLâ1 DNS, 0.4 M NaCl) for 15âmin at 95â°C and cooled to RT before measurement at 540ânm (Infinite 200 PRO M PLEX, Tecan, Männedorf, Switzerland). Values were compared against those of solutions containing only polysaccharide or only enzyme. Statistics of reducing-end assays were performed using a one-way Studentâs t test with an FDR of â¥0.05.
FACE was performed with 8-aminonaphthtalene-1,3,6-trisulfonic acid (ANTS) as fluorophore. 100âμL of the reaction samples were dried in a SpeedVac (Concentrator Plus, Eppendorf) and dissolved in 4âμL 0.05 M ANTS (in DMSO, 15% (v/v) acetic acid) and 4âμL 1 M NaCNBH3 (in DMSO). They were incubated over night at 37â°C before being loaded onto a FACE-Gel86 and separated at 400âV for 1âh.
Thin-layer chromatography
25âμg purified protein were incubated with 0.5% (w/v) poly-/oligosaccharide in PBS for 24âh. Samples were heat-inactivated at 80â°C for 10 min and centrifuged (13,000âÃâg, 10âmin) to remove precipitated protein. Glucose, maltose and maltotriose (all 1âmg mLâ1) in PBS were used as standard for the chromatography. The samples were analyzed as described previously87 on silica gel plates (60 F245) with a mixture of 1-butanol, acetic acid and water (2:1:1) as solvent. Plates were sprayed with staining solution (4âg α-diphenylamine, 4âmL aniline, 200âmL acetone, 30âmL phosphoric acid 80% (v/v)) and visualized by heating above 100â°C.
Affinity gel electrophoresis
Gel electrophoresis was carried out using native 12% acrylamide-gels as described previously88. Purified Flavimarina sp. SusD was diluted with 10âmM Tris-HCL pH 7.4 to decrease the NaCl concentration to 20âmM. Flavimarina sp. SusD and BSA, which was used as non-binding control, were loaded onto gels containing either no additive or 0.2% of the tested polysaccharide. Runs were performed at 80âV with cooled buffer on ice. Gels were stained using Coomassie Brilliant Blue.
Protein structure prediction
Structure models of Flavimarina sp. Hel_I_48 (Supplementary Dataset 5) and Muricauda sp. MAR_2010_75 SusD (Supplementary Dataset 6) were predicted using ColabFold89 using default settings with the top-ranked structure relaxed (num_relaxed: 1). These models were used for an overlay with the structure of the experimentally characterized homolog from Bacteroides thetaiotaomicron in complex with cyclodextrin to determine differences between the SusD binding sites. Models were colored based on the AlphaFold confidence score (pLDDT) and figures were created using PyMOL (Schrödinger, New York, NY, USA)90.
Lysate activity measurements
Samples were taken from cultures growing on 0.1% (w/v) of a single carbon source or 0.2% (w/v) intracellular enriched bacterial extract. Bacteria were harvested by centrifugation (4000âÃâg, 10âmin) and lysed via sonication on ice in PBS (3âÃâ2âmin, 50% cycle). Debris was removed via centrifugation (13,000âÃâg, 10âmin). Supernatant protein concentration was measured by BSA-assay and 25âμg of protein were incubated with 0.5% of tested polysaccharide for 24âh at RT. Samples containing only polysaccharide or only extract were treated similarly as controls. Lysate activity was measured via DNS-Assay and FACE as described above. Significance of the results was determined via one-way ANOVA followed by post hoc Tukeyâs HSD test.
Polysaccharide extraction
Polysaccharides were extracted from enriched intracellular fractions of Polaribacter sp. Hel_I_88. 200âmL culture were harvested via centrifugation (4000âÃâg, 20âmin, 4â°C) and washed once with 10âmL MOPS buffer (20âmM, pH 8) before being resuspended in ddH2O. Polysaccharide extraction was carried out according to a protocol modified from literature91. In short, attached particles were removed by centrifugation (500âÃâg, 10âmin) and cells were lysed by sonication on ice (3âà 2âmin, 50% cycle). Two more centrifugation steps (1100âÃâg, 30âmin and 27,000âÃâg, 15âmin) were carried out to remove unbroken cells and membrane fragments. The pellets were pooled and served as control to ensure enrichment of intracellular polysaccharide (attached fraction). Three volumes of glycine-buffer (0.2 M, pH 10.5) and two volumes of chloroform (both 4â°C) were added to the supernatant and shaken vigorously for 30âs. Phase separation was achieved by centrifugation (100âÃâg, 2âmin) and the aqueous phase removed. The remaining organic phase was re-extracted twice with 2 volumes of glycine buffer and all aqueous phases pooled. They were centrifuged at 47,000âÃâg for 3âh until a gelatinous pellet remained. After resuspending the pellet in 5âmL ddH2O, 6 volumes of ethanol (4â°C) were added to precipitate the polysaccharides over night. Precipitate was then centrifuged (14,000âÃâg, 1âh), resolubilized in ddH2O, dialyzed against ddH2O over night to remove residual salts and finally dried in a SpeedVac. Extracts were weighed and stored at â20â°C until further use.
Bacterial glycan extract characterization
To determine specific components of the bacterial polysaccharide extracts, 5âmg extract were resuspended in PBS and then were incubated with 25âμg of the characterized enzymes GH13A, GH13B, GH13C) and C. forsetii GH1688, respectively. Samples were analyzed by reducing-end assay and FACE as described above. Mono- and oligosaccharide release was compared to samples containing either untreated extract or extract after acid hydrolysis. For acid hydrolysis, 5âmg glycan extract were boiled with 1 M HCl for 2 h, and subsequently neutralized using 1 M NaOH. Monosaccharide composition of all samples was determined via HPAEC-PAD using a Dionex CarboPac PA10 column (Thermo Fisher Scientific) and monosaccharide mixtures as standards92.
Determination of glucan concentrations on filters
Polysaccharide extraction was performed from 3 and 0.2âμm bloom membrane filters of the spring bloom samples and from bacterial single-cultures. Analysis of the extracts was carried out as described93. In short, membrane filters were cut into small pieces and extracted using hot ddH2O with sonication treatment and debris was removed via centrifugation (4500âÃâg, 15âmin). α-glucan content was determined via incubation with amylase (Aspergillus oryzae, Megazyme) and amyloglucosidase (Aspergillus niger, Merck) in sodium acetate buffer (0.1 M, pH 4.5) followed by a PAHBAH-Assay94. The measured values were corrected by the amount of water filtered, thereby taking bacterial cell numbers into account.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.