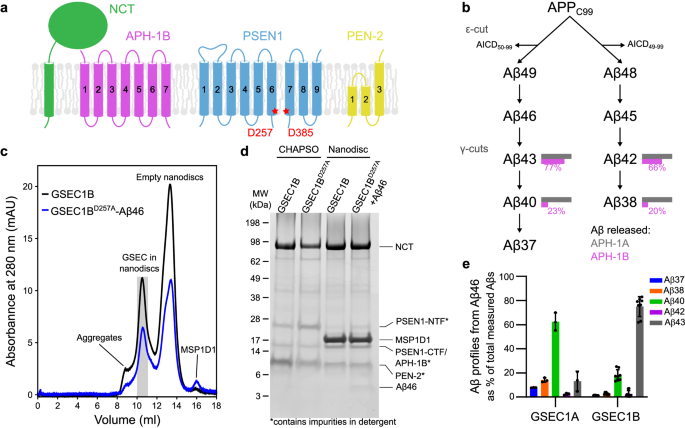
GSEC1B expression, purification and formation of GSEC1B-Aβ46 complex
Human NCT, PSEN1, APH-1B and PEN-2 were expressed in High Five insect cells using a baculovirus expression system as previously described11. NCT was cloned with a PreScission protease cleaving site and GFP tag at the C-terminus. The same system was used to express inactive GSEC1B complex (GSEC1BD257A) in which PSEN1 was expressed as N- and C-terminal fragments (amino acids 1-297 and 298-467, respectively) to mimic the autoproteolytic activation of GSEC.
All the purification steps were carried out at 4â°C. Seventy two hours after infection, cells were collected by centrifugation (4800 x g, 20âmin) and resuspended in 100âml of lysis buffer (25âmM PIPES pH 7.4, 300âmM NaCl, 10% glycerol, 1x Complete protease inhibitor cocktail (Roche)) per litre of culture. Resuspended cells were lysed using Emulsiflex C3 homogeniser (Avestin) and total membrane fractions isolated by ultracentrifugation (100,000 x g, 1âh). Membrane pellets were washed twice in 50âml of high-salt wash buffer (25âmM PIPES pH 7.4, 1âM NaCl, 10% glycerol) per litre of culture; pellets were resuspended using a PTFE plunger in a Heidolph overhead stirrer, incubated on a rotator for 30âminutes and pelleted by ultracentrifugation. Washed membranes were resuspended in solubilisation buffer (25âmM PIPES, 300âmM NaCl, 2% CHAPSO (Anatrace), 5% glycerol, 1 x Complete protease inhibitor cocktail). After overnight incubation at 4â°C, the soluble fraction was separated by ultracentrifugation and incubated overnight with agarose resin NHS-coupled to anti-GFP nanobodies56 (NHS-activated Sepharose 4 FF; Cytiva). Resin was transferred into a gravity column (Bio-Rad) and washed with 20 column volumes (CV) of solubilisation buffer followed by 10 CV of wash buffer (25âmM PIPES, 300âmM NaCl, 1% CHAPSO, 0.1% 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC; Avanti), 5% glycerol) and 10 CV of elution buffer (25âmM PIPES, 150âmM NaCl, 1% CHAPSO, 0.1% POPC, 1âmM dithiothreitol (DTT), 1âmM ethylenediaminetetraacetic acid (EDTA)). Next, the resin was resuspended in 1 CV of elution buffer and GSEC1B was eluted by overnight incubation with 50 μg/ml of PreScission protease57. To remove PreScission protease, the eluted fraction was incubated overnight with Glutathione Sepharose 4B resin (Cytiva). Protein concentration was estimated using Bradford reagent (Bio-Rad) following the manufacturerâs instructions and was in the 0.7â1.5âmg/ml range. Protein purity was assessed by SDS-PAGE (4-12% Bis-Tris; Invitrogen) and Coomassie staining (InstantBlue; Abcam). Purified protein was flash-frozen and stored at â80â°C.
To form GSEC1B-Aβ46 complex, 5 μM Aβ46 (rPeptide) resuspended in dimethyl sulfoxide (DMSO) was added to purified GSEC1BD257A (1.25 x fold excess), followed by a 1âh incubation at 37â°C.
Expression and purification of membrane scaffold protein
Plasmid with MSP1D1, pMSP1D158 (Addgene) was transformed into E. coli BL21(DE3) competent cells. Culture was grown in LB medium containing 25 μg/ml kanamycin at 37â°C until OD600 of 0.8. Protein expression was induced by adding 1âmM IPTG and carried out for 3âh. Cells were collected by centrifugation (4800 x g, 20âmin, 4â°C) and the pellet stored at â20â°C.
All the purification steps were carried out at 4â°C. Cell pellet was resuspended in 50âml of lysis buffer (150âmM Tris pH 8, 300âmM NaCl, 20âmM imidazole pH 8, 2.5âmM MgCl2, 0.1âmM CaCl2) per litre of culture and incubated with a few grains of DNAse I (Sigma) for 1âh. Resuspended cells were lysed using a continuous flow cell disruptor (Constant Systems) and supplemented with 1% Triton X-100. Cell lysate was centrifuged (40,000 x g, 20âmin), and the supernatant was filtered through a 0.45âμm filter before it was applied onto a HisTrap HP (Cytiva) column using an ÃKTA pure system (Cytiva). The column was washed with 10 CV of wash buffer 1 (40âmM Tris pH 8, 300âmM NaCl, 20âmM imidazole pH 8, 1% Triton X-100), 10 CV of wash buffer 2 (40âmM Tris pH 8, 300âmM NaCl, 20âmM imidazole pH 8, 2.15% sodium cholate (Sigma), 1% Triton X-100), 10 CV of wash buffer 3 (40âmM Tris pH 8, 300âmM NaCl, 20âmM imidazole pH 8, 1% sodium cholate), and 10 CV of wash buffer 4 (40âmM Tris pH 8, 300âmM NaCl, 50âmM imidazole pH 8, 1% sodium cholate). MSP1D1 was eluted using an imidazole gradient (from 50 to 400âmM) over 10 CV in wash buffer 4. The peak fractions containing MSP1D1 were pooled and dialysed overnight in 3.5âkDa MWCO cellulose tubing (Carl Roth) against 80 x volume of dialysis buffer (40âmM Tris pH 8, 150âmM NaCl) to remove excess of imidazole. Protein concentration was measured spectrophotometrically (ε280 21430âMâ1cmâ1, MW 24.79âkDa) using NanoDrop One (Thermo Scientific). Dialysed MSP1D1 was concentrated to 5âmg/ml using a 10âkDa MWCO Amicon Ultra centrifugal concentrator (Millipore), supplemented with CHAPSO (4%) and DTT (2âmM) and, to cleave the His-tag, the sample was incubated overnight with TEV protease (1 μg of TEV protease per 100 μg of MSP). The TEV protease was expressed and purified as previously described59. Cleaved MSP1D1 was applied onto a HisTrap HP column (Cytiva), the flowthrough containing untagged MSP1D1 was collected, concentrated to 5âmg/ml, and further purified by size exclusion chromatography using a Superdex 75 column (Cytiva) in SEC buffer (25âmM PIPES pH 7.4, 150âmM NaCl, 0.5% CHAPSO). Protein concentration was measured spectrophotometrically (ε280 18450âMâ1cmâ1, MW 22.04âkDa). Fractions containing pure untagged MSP1D1 were pooled and concentrated to ~10âmg/ml, then flash frozen and stored at -80â°C.
Reconstitution of GSEC1B in lipid nanodiscs
We screened reconstitution conditions including two constructs of membrane scaffold protein (MSP) and two lipid compositions. The optimal reconstitution was obtained using MSP1D1 and POPC:DLPC (1,2-dilauroyl-sn-glycero-3-phosphocholine; Avanti) mix at 1:1 molar ratio.
All the steps were carried out at 4â°C. Purified GSEC1B or GSEC1B-Aβ46 complex at a concentration of 0.7â1.5âmg/ml was diluted with a solution containing 10% CHAPSO and 1.67% DLPC to a final concentration of 1.2âmM POPC and 2.4âmM DLPC and the solution was stirred for 30âmin. MSP1D1 was added to the solution to obtain the MSP:POPC:DLPC molar ratio of 1:60:80 and the solution was stirred for 1âh. Reconstitution of nanodiscs was achieved upon detergent adsorption by Bio-Beads SM-2 resin (Bio-Rad) which was added in three batches, 0.25âg/ml each, and the protein solution was incubated while stirring for 1âh after the first batch, overnight after the second, and 1âh after the third. Nanodiscs were collected from the tube by centrifugation after puncturing the bottom of the tube. The reconstituted GSEC1B in nanodiscs was separated from empty nanodiscs by size exclusion chromatography using a Superdex 200 Increase column (Cytiva) in SEC buffer (10âmM PIPES pH 7.4, 100âmM NaCl). SDS-PAGE and silver staining (Pierce silver stain kit; Thermo Scientific) were used to assess the contents of the fractions.
Peak fractions containing GSEC1B were concentrated to 0.04â0.1âmg/ml using a 100âkDa MWCO Amicon Ultra centrifuge filter (Millipore).
GSEC1B and GSEC1BD257A purified in CHAPSO and reconstituted into nanodiscs were resolved by SDS-PAGE in a 4â12% Bis-Tris NuPAGE gel (ThermoScientific) and transferred to a nitrocellulose membrane. The following primary antibodies and dilutions were used for immunoblotting: anti-NCT (BD Biosciences Cat# 612290, RRID:AB_399607) 1 in 2000, anti-PSEN1NTF (Millipore Cat# MAB1563, RRID:AB_11215630) 1 in 1000, anti-PSEN1CTF (Cell Signalling Technology Cat# 5643, RRID:AB_10706356) 1 in 1000, anti-APH-1B (B78; kindly provided by prof. Bart de Strooper) 1 in 1000, and anti-PEN-2 (Cell Signalling Technology Cat# 8598, RRID:AB_11127393) 1 in 500. The following secondary antibodies were used: goat anti-mouse IgG-HRP conjugate (Bio-Rad Cat# 1721011, RRID:AB_2617113) 1 in 10000, rabbit anti-rat IgG-HRP conjugate (Thermo Fisher Scientific Cat# 61-9520, RRID:AB_2533945) 1 in 2000, and goat anti-rabbit IgG-HRP conjugate (Bio-Rad Cat# 172-1019, RRID:AB_11125143) 1 in 10000. Blots were developed using Western Lightning Plus-ECL Enhanced Chemiluminescence Substrate (Perkin Elmer).
GSEC1B activity assays in nanodiscs
The activity assays were carried out at 37â°C in a Labcycler Gradient thermocycler (SensoQuest). Final buffer composition of the reactions was 25âmM PIPES, 150âmM NaCl, 0.025% DMSO. GSEC1A was expressed, purified, and reconstituted into nanodiscs as described above. Assays were carried out with 0.23 μM GSEC1A/GSEC1B/GSEC1BD257A and 2.5 μM Aβ46 for 24âh. Reactions were quenched by placing the assay tubes on ice and adding 10 μM inhibitor L-685,458 (Santa Cruz Biotechnology). De novo Aβ (37, 38, 40, 42) production was quantified by MSD ELISA as described previously52, and Aβ43 was quantified using Amyloid-beta (1-43) (FL) ELISA kit (IBL) according to the manufacturerâs instructions. The plots were generated using GraphPad Prism version 8.4.2.
Generation of MEF cell lines and the cell-based activity assays
Single-point mutations (Y115A, Y115F, W165F and S169A) were introduced into human PSEN1 cDNA and cloned into the pMSCVpuro vector using Q5 Site-Directed Mutagenesis Kit (NEB BioLabs) according to the standard protocol. The sequences of the primers used are available in Supplementary Data 1. To generate recombinant retroviruses, the generated vectors and the PIK packaging plasmid were delivered into HEK 293âT cells using the TransIT-LT1 transfection reagent (Mirus Bio). Stable MEF cell lines expressing either WT or mutant PSEN1/GSEC complexes were generated through retroviral transduction of Psen1â/â/Psen2â/â mouse embryonic fibroblasts (MEFs)60 as previously described52. MEFs were cultured in Dulbeccoâs Modified Eagleâs Medium (DMEM)/F-12 (Life Technologies) supplemented with 10% foetal bovine serum (FBS) (Sigma-Aldrich)35. Cell lines stably expressing the WT/mutant proteins were selected using media supplemented with 5 μg/ml puromycin (Sigma-Aldrich). The reconstitution of mutant GSECs was assessed by SDS-PAGE and western blot. Briefly, cells were collected, and total membranes isolated and solubilised in 28âmM PIPES pH 7.4, 210âmM NaCl, 280âmM sucrose, 1.5âmM EGTA pH 8, 1% CHAPSO, 1x Complete protease inhibitor cocktail. Equal amounts of solubilised protein were resolved in a 4â12% Bis-Tris NuPAGE gel (ThermoScientific) and transferred to a nitrocellulose membrane. The following primary antibodies were used: anti-NCT (9C3; kindly provided by Prof. Wim Annaert) 1 in 2000, anti-PSEN1CTF (as described above), and anti-PEN2 (as described above). Secondary antibodies used and blot development was done as described above.
To assess the effects of PSEN1 mutants on Aβ production, APPC99 substrate was transiently expressed in the generated MEF cell lines using a recombinant adenoviral expression system as previously described61. In brief, cells were plated in a 96-well plate at the density of 12,500 cells/well and transduced 6-8âh later with Ad5/CMV-APPC99 adenovirus52. The culture medium was replaced with a low-serum medium (DMEM/F-12 medium containing 0.2% FBS) 16âh later, and collected after a 24âh incubation period at 37â°C.
The conditioned media was cleared by centrifugation at 800 x g for 15âmin and used to determine Aβ (37, 38, 40, 42) peptide levels using MSD ELISA as described previously52, and Aβ43 was quantified using Amyloid-beta (1-43) (FL) ELISA kit (IBL) according to the manufacturerâs instructions. The plots were generated using GraphPad Prism version 8.4.2.
Preparation of graphene oxide coated EM grids
Carbon-coated holey grids Quantifoil R 0.6/1 Cu 300 and CF-2/1-3âC (EMS) were used for preparing cryo-EM samples for apo GSEC1B and GSEC1B-Aβ46 complex, respectively. Graphene oxide 0.2% (w/v; Sigma) and 1% (w/v; GOgraphene) were used interchangeably.
Grids were glow-discharged with carbon face up using ELMO glow discharge system (Agar Scientific) with 5âmA current for 1âmin at 0.3 mbar in air. A volume of 4 μl of 0.5âmg/ml poly-L-lysine solution (MW 15â30âkDa; Sigma) in 10âmM PIPES pH 7.4, 100âmM NaCl was pipetted on the carbon side of the grid and incubated for 2âmin, blotted with Whatman grade 2 paper and washed twice by pipetting 4âµl of MilliQ water on the carbon side, followed by blotting. After 5âmin drying on air, 3 μl of 0.2âmg/ml graphene oxide was pipetted on carbon side of the grid and incubated for 2âmin. Next, the grid was blotted, and washed three times by touching a droplet of water with the GO side, followed by blotting. The grid was dried for 30âmin and 4 μl of 0.1% PEG 10,000 (Fluka Chemica) in 10âmM PIPES pH 7.4 and 100âmM NaCl was applied on the carbon side of the grid and incubated for 2âmin. The grid was blotted and washed twice by pipetting 4 μl of water on top and blotting. The grid was dried for 5âmin and used for preparation of cryo-EM samples immediately.
Cryo-EM sample preparation
GSEC1B nanodisc solution (2 μl) at concentration of 0.04âmg/ml was pipetted on the front side of graphene-oxide-coated (âMRC protocolâ62) Quantifoil R 0.6/1 Cu300 grid and incubated for 15-30âsec at 99% humidity in Cryoplunge 3 (Gatan). The grid was blotted from both sides for 2.5-3âsec with Whatman grade 3 paper and plunged into liquid ethane at -175â°C.
For the GSEC1B-Aβ46 complex, 5 μM Aβ46 (15-fold excess) resuspended in DMSO was added to GSEC1B-Aβ46 reconstituted into lipid nanodiscs (1% final DMSO concentration) and the mix was incubated for 1âh at 37â°C. Next, the protein solution was cooled on ice and plunge frozen on CF-2/1-3âC grids coated with graphene oxide (according to the protocol described in the section above) as described for the apo protein complex.
Cryo-EM data collection
The micrographs were collected on a CRYO ARM 300 electron cryogenic microscope (JEOL) equipped with an in-column Omega energy filter and operated at 300âkeV. The energy filter slit was set to 20âeV and the data were collected at a nominal magnification of 60,000 with a magnified pixel size of 0.766âà on a K3 direct electron detector (Gatan). SerialEM (3.8.2 for apo, 3.8.18 for Aβ46-bound data) was used for automated data collection.
The micrographs for GSEC1B in apo state were collected as 60-frame movies with an electron dose of 1.06 eâ à â2 per frame over 3âseconds in a 5âÃâ5 pattern with one exposure per 0.6âμm nominal and 0.3âμm measured hole diameter, producing 25 micrographs per stage position. A total of 10,733 movies were collected from one EM grid with the defocus in the range from â1.2 to â2 μm.
For the GSEC1B-Aβ46 complex the micrographs were collected in a 3âÃâ3 pattern with 5 exposures per each 2âμm hole resulting in 45 movies per stage position. Each movie contained 60 frames with total exposure of 2.8âs and an electron dose of 0.936 e- à â2 per frame. A total of 18,855 movies were collected. The defocus was set to â1.3 to â2.3âμm.
Image processing
Movies were pre-processed on-the-fly using RELION 3.1 schedules63 as a wrapper to run MotionCor2 1.4.264 for frame alignment and CTF parameters were estimated using CTFFIND 4.1.1465. Further processing was done using RELION 3.1 unless otherwise stated. Particles from a subset of micrographs were auto-picked using crYOLO 1.766 with the general model and submitted to 2D classification in either RELION or cryoSPARC (v3.2.0 for the apo dataset, v3.3.1 for the Aβ46-bound dataset)67. 100 micrographs containing the highest number of particles from good classes were manually screened to remove bad particles, then these micrographs and particle coordinates were used to refine the crYOLO general picking model. The refined model was used to auto-pick the entire dataset.
For the apo dataset, 986,830 particles were picked from 5501 micrographs and extracted in a 320âÃâ320 pixel box downsampled to 64âÃâ64 pixels (Supplementary Fig. 3). 2D classification was done in RELION (ignoring CTFs until the first peak) and in cryoSPARC (setting initial classification uncertainty factor to 20, online-EM iterations to 100, final full iterations to 20, batch size per class to 1000, enforcing non-negativity, activating clamp-solvent option, disabling FRC-based regularizer, full FRC, setting iteration to start annealing sigma to 10, number of iterations to anneal sigma to 50, and using white noise model). Particles belonging to good 2D classes from both programs were merged and duplicate particles removed resulting in a total of 790,832 particles which were then reextracted in a 320âÃâ320 pixel box downscaled to 128âÃâ128 pixels. Ab-initio reconstruction in cryoSPARC was used to generate an initial model from a subset of particles. 3D classification with 1 class and using tau_fudge value of 64 was used to centre and align the particles. 3D classification with 4 classes and tau_fudge value of 64 resulted in 2 classes with visible transmembrane helices accounting for 413,321 particles which were then reextracted in a 320âÃâ320 pixel box downscaled to 256âÃâ256 pixels. 3D auto-refinement of these particles resulted in a map resolved to an average resolution of 3.8âà . Next, 3D classification without alignment with 2 classes and tau_fudge value of 8 was used to separate particles contributing to high-resolution reconstruction and resulted in 115,197 particles. A 3.6âà map was reconstructed from these particles which was improved to 3.3âà after reextracing in a 320âÃâ320 pixel box without downsampling, Bayesian polishing and defocus refinement. To better resolve the transmembrane region, another round of 3D refinement was performed using external reconstruction with SIDESPLITTER68 which resulted in the final map at a resolution of 3.2âà . The final map was sharpened using post process procedure in RELION. The pixel size was calibrated using the NCT ectodomain from the GSEC1A-C83 structure (PDB: 6IYC) as a reference in UCSF Chimera69, by fitting the reference model into the experimental map. The final map was rescaled and sharpened using the calibrated pixel size of 0.776âà yielding a 3.3âà map. The map was further filtered using local resolution filter and the B-factor determined during post processing. The local filtered map was used for model building and deposited to EMDB.
For the GSEC1B-Aβ46 complex, 2,788,683 particles were picked from 18,855 micrographs and classified in 2D as described above (Supplementary Fig. 5). 2,433,778 particles selected after 2D classification were centred and aligned using a 3D classification with 1 class and using tau_fudge value of 64. 3D classification with 10 classes, tau_fudge value 64 over 75 iterations was done using a low-pass filtered map from the apo dataset as a reference model. 3D classes with visible transmembrane helices from iterations 51-75 were picked and duplicate particles removed for a total of 2,023,687 particles which were then subjected to 3D refinement to obtain a map resolved to 4.1âà . The densities for PSEN1 TM2 and Aβ46 were very weak. Further 3D classification of these particles (Kâ=â10; Tâ=â4; 3.7° sampling; 15° search range; 25 iterations) resulted in two high-resolution classes containing 397,175 particles which were subjected to 3D refinement and resulted in a 3.5âà map with improved, but still weak densities of PSEN1 TM2 and Aβ46. CTF refinement including anisotropic magnification, beam tilt, trefoil, per particle defocus and per micrograph astigmatism followed by Bayesian polishing improved the resolution of the reconstruction to 3.3âà . A mask containing PSEN1 and Aβ46 density was used to subtract the rest of the signal and the subtracted particles were subjected to 3D classification without alignment (Kâ=â10; Tâ=â32; 400 iterations) which resulted in a class containing 53,612 particles with improved densities of PSEN1 TM2 and Aβ46. These particles were reverted and subjected to 3D refinement which yielded a map resolved to 3.4âà . The pixel size was calibrated to 0.949 (320âÃâ320 pixel box downscaled to 256 pixels) and the map was postprocessed, sharpened and filtered in the same way as described above, yielding a 3.4âà map. The local filtered map was used for model building and deposited to EMDB.
Model building and refinement
For the apo structure, the starting model was obtained from the structure of GSEC1A in amphipols (PDB: 5FN5)28 and rigid-body-fitted into the EM density using UCSF Chimera69. A model of APH-1B was built with by homology modelling using modeller 970. The model was further manually built and refined using Coot 0.9.871 after which it was refined using real-space refinement with simulated annealing in PHENIX 1.19.272. Several iterations of manual rebuilding in Coot and automated refinement in PHENIX with secondary structure restraints were used to refine the model. The model was validated using MolProbity73.
For the GSEC1B-Aβ46 structure, the starting model was obtained from the structure of GSEC1A-C83 (PDB: 6IYC)20 from which the sugars, substrate and lipids were removed. Aβ46 was manually built as a polyalanine model. PSEN1 loop 1 (amino acids 106-123) was manually built to fit into the density and the resulting PSEN1 model was used for template-based structure prediction using ColabFold74,75 to aid manual building of loop 1. Further refinement and validation were done as described above.
To make structural comparison possible, the pixel size of the apo GSEC1A structure (EMDB-3240)28 was calibrated and the map was rescaled in the same way as described above. To avoid reinterpreting the map, the model (PDB: 5FN5) was refined into the rescaled map using real-space refinement in PHENIX and the original model was fitted onto the backbone of the refined model five amino acids at a time with an overlap of two amino acids to keep the sidechain orientations intact.
Figures of atomic models and cryo-EM maps were generated using UCSF ChimeraX76 version 1.4.
Bioinformatics analysis
APH-1 sequences were aligned using Clustal Omega77 and coloured using Jalview78 version 2.11.2.6 according to BLOSUM62 score with a 40% conservation threshold.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.