Reagents and materials
All the chemicals were of analytical grade or of higher purity and were purchased from Sigma‒Aldrich, Macklin (Shanghai, China), and Sinopharm (Beijing, China) unless otherwise noted. Primers and genes were synthesized by Tsingke (Beijing, China), and Primers were dissolved in Milli-Q water to a final concentration of 10 μM. PrimeSTAR Max DNA Polymerase was obtained from Takara Bio (Japan). The methylation-sensitive restriction enzyme (DpnI) was obtained from New England Biolabs. A ClonExpress-II one-step cloning kit was obtained from Vazyme (Nanjing, China). Lysogeny Broth (LB) medium (1 L) containing 10 g of NaCl, 10 g of tryptone and 5 g of yeast extract. For the LB agar plates, 20 g of agar was added. All the media were then autoclaved at 121 °C for 20 min. For the anaerobic culture medium, 100 mM 3-(N-morpholino) propanesulfonic acid (MOPS), 5 g L−1 glucose, 25 mM sodium fumarate, and 1 mg L−1 resazurin were added to LB media, and the pH was adjusted to 7.4. The anaerobic culture medium was autoclaved at 115 °C for 30 min, after which the oxygen was replaced with 100% N2. Carbon nanotubes (CNTs) were obtained from XFNANO (Nanjing, China).
Construction of plasmids
The hdcr gene was codon optimized and synthesized and subsequently cloned and inserted into pET20b by Tsingke. The polymerase chain reaction (PCR) mixture (50 μL) consisted of 25 μL of PrimeSTAR Max DNA Polymerase, 20 ng of the template (pET20b-hdcr), and 500 nM primers P1 and P3, followed by the PCR protocol: 98 °C for 2 min (1 cycle); 98 °C for 20 s; 53 °C for 20 s; 72 °C for 2 min (25 cycles); and 72 °C for 10 min (1 cycle). PCR products were characterized by agarose gel electrophoresis. The Megawhop PCR mixture (50 μL) contained 25 μL of PrimeSTAR Max DNA Polymerase, 20 ng of the pTrcHisA plasmid as the template, and 500 nM primers P2 and P4. The PCR protocol was as follows: 98 °C for 2 min (1 cycle); 98 °C for 30 s, 53 °C for 20 s, and 72 °C for 2 min (25 cycles); and 72 °C for 10 min (1 cycle). All the methylated templates were specifically degraded by DpnI. The PCR products were purified with a universal DNA purification and recovery kit (TIANGEN, China, Beijing). The above two PCR products were linked with a ClonExpress-II one-step cloning kit (37 °C, 30 min). The linked product was subsequently transformed into 100 μL of E. coli Top10 competent cells (heated at 42 °C for 45 sec) and recovered at 37 °C for 1 h. The cells were subsequently plated on LB agar plates supplemented with 50 mg L−1 ampicillin for growth overnight at 37 °C.
The construction of the plasmid pTrc99a was performed with kanamycin resistance. The kanamycin resistance gene was cloned from the template pET28a using the primers P11 and P12. pTrc99a was generated by removing the original ampicillin resistance gene via the primers P13 and P14, and the pTrc99a-kan+ plasmid harboring the kanamycin resistance gene was generated via the ClonExpress-II one-step cloning kit. The linked product was subsequently transformed into 100 μL of E. coli Top10 competent cells, which were subsequently allowed to recover at 37 °C for 1 h. The cells were plated on LB agar plates supplemented with 50 mg L−1 kanamycin for growth overnight at 37 °C.
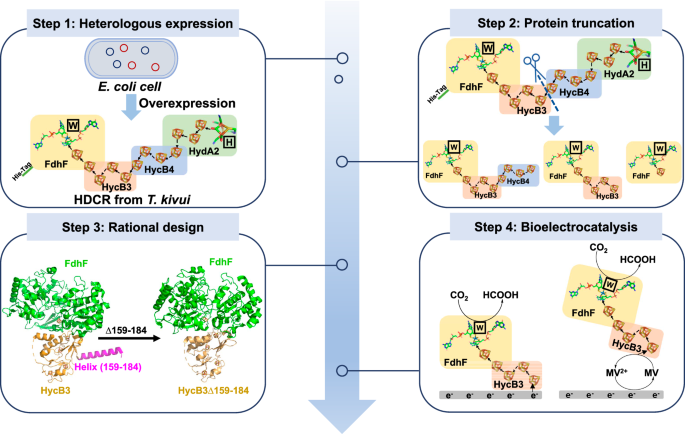
The plasmid pTrc99a-kan+–hydGXEF containing the maturation factor genes hydG, hydX, hydE, and hydF of FeFe-hydrogenase from Shewanella oneidensis MR-1 was constructed using the primers P15, P16, P17 and P18. pTrc99a-kan+–hydGXEF and pTrcHisA-hdcr were subsequently transformed into E. coli MC1061 competent cells. The clone was picked from LB agar plates containing kanamycin and ampicillin. E. coli MC1061 containing pTrc99a-kan+–hydGXEF and pTrcHisA-hdcr was used to express HDCR. The pTrcHisA-fdhF_hycB3_hycB4 (pTrcHisA-hdcr∆hydA2), pTrcHisA-fdhF_hycB3 (pTrcHisA-hdcr∆hydA2∆hycB4), and pTrcHisA-fdhF (pTrcHisA-hdcr∆hydA2∆hycB4∆hycB3) were constructed using primers P5 and P6 P5 and P7, and P5 and P8, respectively. The plasmid was transformed into E. coli MC1061 competent cells. The clone was picked from the LB agar plates containing ampicillin. The FdhF_HycB3∆159-184 variant-expressing plasmid pTrcHisA-fdhF_hycB3∆159-184 (pTrcHisA-hdcr∆hydA2∆hycB4∆159-184) was constructed by using pTrcHisA-fdhF_hycb3 as a template, and using primer P9 and P10. The variant FdhF_HycB3-C83S expression plasmid was constructed by using primer P38 and P39, and pTrcHisA-fdhF_hycB3 was used as the template.
The plasmid pETDuet-hdcr was constructed by cloning fdhF and hycb3 into multiple cloning sites 1 (MCS1) of pETDuet-1 using P19 and P20, P21 and P22. The hycB4 and hydA2 genes were subsequently cloned and inserted into the MCS2 of pETDuet-1 using P23 and P24, P25, and P26. The maturation factor gene FdhD of FdhF was also cloned and inserted into MCS1 of pETDuet-1 using P20 P27, P28, and P29. In addition, the pACYCDuet-hydGXEF plasmid, which contains the maturation factor genes hydG, hydX, hydE, and hydF of FeFe-hydrogenase from Shewanella oneidensis MR-1, was constructed by cloning the hydG and hydX genes into the MCS1 of pACYCDuet-1 using P30 and P31, P32 and P33, and cloning hydE and hydF into the MCS2 of pACYCDuet-1 using primers P34 and P35, P36 and P37. Both pETDuet-hdcr and pACYCDuet-hydGXEF were transformed together into E. coli BL21 (DE3) competent cells. The clone was picked from LB agar plates containing ampicillin and chloramphenicol. For primer sequences, please refer to Supplementary Table 1.
Protein expression and purification
For the expression of HDCR in E. coli BL21 (DE3), cells were grown anaerobically in anaerobic culture medium29 supplemented with ampicillin (50 mg L−1), chloramphenicol (30 mg L−1), and 1 mM sodium tungstate. Cultures were grown anaerobically at 37 °C to an OD600 of 0.6–0.8, 2 mM FeSO4, and 0.4 g L−1 L-cysteine were added. A final concentration of 0.2 mM isopropyl beta-D-1-thiogalactopyranoside (IPTG) was used for the induction of protein production for 20 h under anaerobic conditions at 18 °C and 150 rpm. For the expression of HDCR in E. coli MC1061, the conditions were the same as above, except for the change from ampicillin (50 mg L−1) or chloramphenicol (30 g L−1) to ampicillin (50 mg L−1) and kanamycin (50 mg L−1). The expression of the HDCR variants in E. coli MC1061 was performed under the same conditions but with only the addition of ampicillin (50 mg L−1).
All purification steps were performed under strictly anaerobic conditions at room temperature in an anaerobic chamber (Vigor Gas Purification Technologies Company, China Suzhou) filled with 96–98% N2 and 2–4% H2, and an oxygen content below 2 ppm. For HDCR purification, cultures were centrifuged at 4 °C for 10 min at 4500 × g, resuspended in 30 mL of buffer A (25 mM Tris, 20% [v/v] glycerol, 20 mM MgSO4, pH 7.5, and 1 mg L−1 resazurin), centrifuged again and resuspended in buffer A supplemented with 40 mg L−1 PMSF, 0.1 g L−1 DNaseI, 2 g L−1 lysozyme, and 0.5 mM dithiothreitol, and then incubated at 37 °C for 1 h. The cells are completely broken up by freezing and thawing three times with liquid nitrogen. Cell debris was removed by centrifugation at 17500 × g and 4 °C for 30 min. The cleared lysate was added to 3 mL of Ni-nitrilotriacetate (Ni-NTA), which was equilibrated with 15 mL of buffer A containing 40 mg L−1 PMSF and 0.5 mM dithiothreitol. The resin was subsequently washed with 25 column volumes of buffer A containing 40 mM imidazole, 40 mg L−1 PMSF, and 0.5 mM dithiothreitol, and the retained protein was eluted with ~ 5 mL of the same buffer A containing 250 mM imidazole, 40 mg L−1 PMSF and 2 mM dithiothreitol. For the purification of the other HDCR variants, buffer B (100 mM Tris, 100 mM NaCl, 10 mM NaNO3, pH 8.0, and 1 mg L−1 resazurin) was used, and the purification procedure was the same as above. The protein purifier (AKTA purifier 10) equipped with Superdex 200 (10/300 GL, General Electric Company) was used for further purification. The flow of buffer was 0.9 mL min−1. The protein concentration was measured according to the Bradford method. The proteins were separated on 15% polyacrylamide gels and stained with Coomassie brilliant blue G250.
Enzyme activity assay
Enzyme activity assays were performed in an anaerobic chamber. Buffer C (HEPES/NaOH 100 mM, 20 mM MgSO4, pH 7.0, 2 mM DTT) was used for HDCR, buffer D (HEPES/NaOH 100 mM, pH 7.0, 2 mM DTT) was used for HDCR variants, and DTT was used as the protective reagent for enzymes. For formate oxidation, 20 mM substrate, sodium formate, and 20 mM electron acceptor, MV2+, were added into Buffer C/D, and the total volume was 2 mL. The absorption at 604 nm (ε = 13.9 mM−1 cm−1), assigned to the reduced MV+, was monitored by Evolution One (Thermo Fisher Scientific) with a temperature control module after the addition of HDCR and its variants. One unit of oxidation activity was defined as oxidizing 1 μmol of formate (reducing 2 μmol of MV2+) per minute under the assay conditions.
For CO2 reduction, MV was first reduced by sodium dithionite (DTH) to produce the reduced MV, which is used as the electron donor for CO2 reduction. Similarly, the absorption decreased at 604 nm (ε = 13.9 mM−1 cm−1), and Evolution One (Thermo Fisher Scientific) was monitored with a temperature control module after the addition of HDCR and its variants. One unit of reduction activity was defined as reducing 1 μmol of CO2 (oxidizing 2 μmol of MV+) per minute under the assay conditions.
For a temperature-dependent experiment, the temperature is controlled by a water bath and temperature-controlling module in a UV-Vis spectrophotometer (Evolution One, Thermo Fisher Scientific). Enzyme aliquots were stored in a water bath at a specific temperature for 10 min, and the pre-mixed reaction buffer without enzyme was incubated inside the spectrophotometer with the temperature module controlled via the software.
Enzyme catalytic CO2 reduction in solution
The enzyme catalytic reaction was performed in an anaerobic chamber. 20 mM of reduced MV was used as the electron donor, CO2 was used as the substrate, and 50 μg of FdhF_HycB3∆159–184 was used as the catalyst in 5 mL of the reaction solution. The control used inactive FdhF_HycB3∆159-184, which was prepared by boiling for 20 min. After 1.5 h at 30 °C, 100 µL samples were taken, and 5 mM sulfuric acid was added to stop the reaction. The samples were centrifuged at 12000 × g for 10 min, and the produced formic acid was detected by high-performance liquid chromatography (HPLC) equipped with a RID−20A detector (Nexera XR, Shimadzu) and an Aminex HPX‐87H column (300 × 7.8 mm, Bio‐Rad). The mobile phase was 5 mM sulfuric acid, the flow rate was 0.6 mL min−1, and the column temperature was 60 °C.
Electrochemical experiments
Electrochemical measurements were carried out using a potentiostat (CHI660e) and a rotating disk electrode (PINE) in the anaerobic chamber with 100% N2. A three‐electrode system was used. The glassy carbon electrodes (0.07 cm2) were polished with an alumina slurry (0.05 μm) on a polishing cloth and then sonicated for 1 min in deionized water and ethanol, respectively. A homogenous black CNT was dispersed in water and dimethylformamide (DMF) (ratio of 1:1) to a concentration of 1 g L−1. A 10 μL aliquot of the CNT suspension was drop-cast on a fresh GC electrode, denoted as the CNT/GC electrode. For the bioelectrode, 10 μL of enzyme (1 g L−1) was drop-cast on the CNT/GC electrode and then left to stand at 4 °C for 4 h in an anaerobic chamber. The excess enzyme solution was gently washed off before testing. A platinum wire was used as the counter electrode, and a saturated Ag/AgCl electrode was used as the reference electrode. All applied potentials were converted versus the standard hydrogen electrode (SHE) using the following equation 1 at 25 °C and pH 7.0.
$${ESHE}={EAg}/{AgCl}+197{mV}$$
(1)
Cyclic voltammetry (CV) and linear sweep voltammetry (LSV) analyses were performed at 25 °C in 100 mM HEPES/NaOH and 100 NaCl at pH 7.0. In the pH-dependence experiment, pH was controlled by using a mixed-buffer system [100 mM Acetate, 2-morpholinoethane sulfonic acid (MES), N-cyclohexyl-2 aminoethane sulfonic acid (HEPES), and N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid (TAPS)], and checked following experimentation. The electrode speed was 900 rpm, the scan rate was 5 mV s−1for CV, and the scan rate was 1 mV s−1 for LSV. The electrolytic cell with a heating layer was used for the temperature optimization experiment. The temperature of the pool is controlled by a flowing water bath, and cyclic voltammetry is performed after reaching the desired temperature, as confirmed by a thermometer test.
Enzyme-electrocatalytic CO2 reduction to formate
A carbon paper (TORAY, Japan) with an area of 1 cm2 was used as the supporting enzyme, 100 μL of CNTs (1 g L−1) was added to the carbon paper, and after drying, 95 μL of FdhF_HycB3∆159-184 solution (1 g L−1) and 5 μL of Nafion (0.02%) were mixed and added to the CNTs to build FdhF_HycB3∆159-184/carbon nanotube/carbon paper (FdhF_HycB3∆159-184/CNT/CP) electrode, after which the mixture was allowed to stand at 4 °C for 4 h in an anaerobic chamber. The excess enzyme solution was gently removed. A three-electrode electrochemical cell with FdhF_HycB3∆159-184/CNT/CP electrode as the working electrode, Ag/AgCl electrode as the reference electrode, and platinum sheet (1 cm2) as the counter electrode was used. Chronoamperometry was carried out in 5 mL of buffer (100 mM HEPES/NaOH, 100 mM NaCl, pH 7.0) that was continuously fed pure CO2, and a constant voltage of − 0.5 V vs. SHE was used. For the MET enzyme-electrocatalytic CO2 reduction to formate, 0.2 mM MV was added to the electrolyte. The formate concentration in the electrolyte was detected by a high-performance liquid chromatography (HPLC) instrument equipped with a RID-20A detector (Nexera XR, Shimadzu) and an Aminex HPX‐87H column (300 × 7.8 mm, Bio‐Rad). The samples were prepared by adding H2SO4 at a final concentration of 5 mM and were centrifuged for 10 min at 12000 × g. The mobile phase was 5 mM sulfuric acid, the flow rate was 0.6 mL min−1, and the column temperature was 60 °C. The standard curve for formate was generated by employing known concentrations of formate ranging from 1 to 100 mM. For the 1H-NMR experiment, 400 μL of electrolyte and 200 μL of D2O were mixed in a clean NMR tube and subsequently analyzed via one-dimensional 1H liquid NMR spectroscopy (Bruker Advance 600 MHZ). The faraday efficiency (FE) and half-cell cathodic energy efficiency (CEE%) of formate were calculated according to Eqs. 2 and 346.
$${FE}\left(\%\right)=\frac{Q({formate})}{Q({tot})}*100\%=\frac{c({formate})*V*Z*F}{j*t}$$
(2)
where c (formate) is the formate concentration after reaction time t (s), V is the volume of the electrolyte, z is the number of electrons required to reduce CO2 to formate, and the value is 2, F is the Faraday constant (96,485 C mol− 1), and j is the recorded current.
$${CEE}\left(\%\right)=\frac{1.23-E(\;{formate})}{1.23-E({applied})}*{FE}*100\%$$
(3)
where E (formate) = − 0.42 V vs. SHE is the reduction potential of CO2 reduction to formate at pH 7.0. FE is the faradaic efficiency of formate production. E (applied) = − 0.5 V vs. SHE is the applied potential vs. SHE.
Dynamic light-scattering assay
A NanoBrook 90plus instrument (Brookhaven Instruments Corporation, USA) was used to analyze the particle size of the proteins. The concentration of all the samples was 0.5 g L−1 and the samples were centrifuged for 10 min at 12000 × g and 4 °C. Each measurement was made three times at 25 °C.
ICP-OES analysis
The W element in FdhF_HycB3∆159-184 was analyzed through ICP-OES (Thermo, ICPOES7200) by Beijing Zhongke Baice Technology Service Co., Ltd.
Bioinformatic methods
All DNA sequences were retrieved from the National Center for Biotechnology Information database, and all protein structures were obtained from the Protein Data Bank. The plasmid map was generated by SnapGene, and the protein structures were visualized using PyMOL. Protein docking analysis was performed via ZDOCK47.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.