Our research complies with all relevant ethical regulations. All animal experiments were carried out according to the German animal welfare act considering the guidelines of the National Institute of Health and the 2010/63/EU Directive of the European Parliament on the protection of animals used for scientific purposes. Animal experiments were conducted with the approval of the Landesamt fûr Gesundheit und Soziales (LAGeSo, Berlin) and the Max-Delbrück-Center for molecular medicine. hiPSC were commercially sourced (GM25256) and were not genetically edited, and as such are not considered relevant material under the 2004 UK Human Tissue Act and their use did not require specific ethical approval.
Cell culture
HEK293 were HEK-tsA201 cells (Sigma-Aldrich ECACC Cat#96121229). HEK293AD cells were from Biocat, cat. no. AD-100-GVO-CB. Gαs-KO cell line were derived from HEK293A from Thermo Fisher Scientific. HEK293 derived cells (Epac-SH187) were derived from HEK-tsA201. Cells were cultured in DMEM medium containing 4.5 g/l glucose (Gibco) supplemented with 10% fetal calf serum (FSC, Biochrome), 2 mM L-glutamine, 100 units/ml penicillin and 0.1 mg/ml streptomycin, at 37 °C, 5% CO2. To split cells, growth medium was removed by aspiration and cells were washed once with 10 mL of PBS (Sigma), followed by trypsinization for 2 min in 1.5 mL of trypsin 0.05%/EDTA 0.02% (PAN Biotech) solution and resuspended in the desired amount of DMEM medium.
Transfection
For the single cell microscopy experiments the cells were transfected directly in the imaging dishes 24 hours prior to conducting the experiments, using JetPrime transfection reagent (Polyplus) according to manufacturer’s protocol. For the plate reader experiments the cells were transfected on 10 cm plates 48 hours before the experiment using a reduced amount of JetPrime transfection reagent and total DNA (5 µl and 2.5 µg respectively) and transferred to black 96-well plates (Brandt) 24 hours before the experiment. The transfected cDNA and cDNA ratios for co-transfections are listed in further sections.
Molecular cloning
The tricistronic Gs and Gs-CFP plasmids were created using NEBuilder HiFi DNA assembly cloning kit (NEB). Gαs (or Gαs-CFP54), Gβ1 and Gγ2 were cloned into the pcDNA3.1 backbone following the same structure as for the published Gi FRET sensors24: Gβ1−2A-Gγ2-IRES-Gαs (or Gαs-CFP respectively). The primers used to generate these constructs are available in Supplementary Table 1.
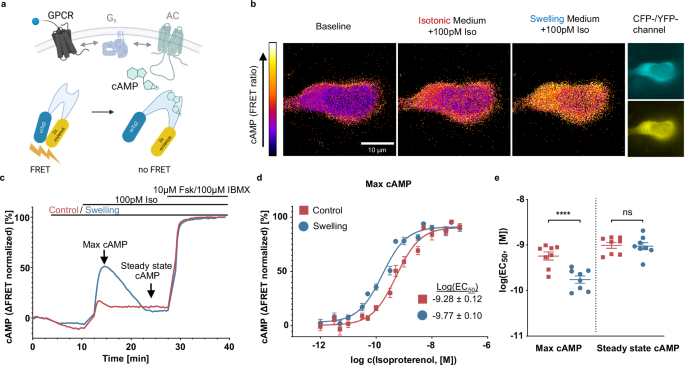
Swelling medium
Throughout all experiments two types of imaging buffers (control – mannitol electrolyte solution 300 mOsm (MES300) and swelling – mannitol electrolyte solution 200 mOsm (MES200)) were used. A basic buffer with a low osmolarity – ES150 – was produced by solving 60 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES in distilled water and resulting in electrolyte solution 150 mOsm (ES150). It was then supplemented with 150 mM mannitol (Sigma) to produce MES300 and reach 300 mOsm. MES300 was then used for washing and incubation steps as described in specific experiments. To achieve hypotonic conditions of 200 mOsm during the experiments, 2/3 of the total volume of MES300 was removed from the wells and replaced with the same volume of ES150, resulting in MES200. In control condition 2/3 of MES300 were also removed and replaced with fresh MES300 to avoid ascribing any effects to mechanical artifacts.
Cardiomyocyte isolation
Cardiomyocyte isolation involved obtaining cardiac myocytes from adult ventricles of seven (7) CAG-Epac1-camps mice aged 8 to 12 weeks9, including 6 males and 1 female. The CAG-Epac1-camps mice (mus musculus, FVB/N transgenic mice) were a kind gift of Kristina Lorenz, Institute for Pharmacology and Toxicology, University of Würzburg. All animal experiments were carried out according to the German animal welfare act considering the guidelines of the National Institute of Health and the 2010/63/EU Directive of the European Parliament on the protection of animals used for scientific purposes. Animal experiments were conducted with the approval of the Landesamt fûr Gesundheit und Soziales (LAGeSo, Berlin) under the internal sacrification license X9010/18, under the institute (MDC) licence X9014/11. The animals had free access to food and water and were kept in individually ventilated cages under a 12 h:12 h light/dark regime (light from 6:30 am to 6:30 pm), a constant 22 ± 2 °C temperature and 55 ± 10% humidity.
The process employed enzymatic collagen digestion and retrograde perfusion through the aorta using a Langendorff perfusion apparatus. In brief, the hearts were swiftly removed from the mice after cervical dislocation and connected to a custom-built perfusion system. Initially, the hearts were perfused with perfusion buffer for 4 minutes at a rate of 3 ml/min, followed by an 8-minute perfusion with myocyte digestion buffer containing 5 mg of liberase dispase high DH enzyme (Roche) per digestion. Subsequently, the heart was detached from the perfusion system, and the ventricles were dissected into small pieces using scalpels. After sedimentation, removal of the supernatant, and resuspension the cells were filtered through a nylon mesh cell strainer (100 µm pore size, Falcon) to eliminate any remaining tissue fragments. The cells were then gradually exposed to physiological Ca2+ concentrations (~1 mM), resuspended in myocyte plating medium, and seeded on freshly coated Matrigel-coated 8-well Ibidi µ-slides for each experiment. More detailed information about the buffers and materials used can be found in the methods section of Bathe-Peters et al9.
hiPSC-CM handling and differentiation
hiPSC-derived cardiomyocytes (hiPSC-CM) were generated according to the protocol previously described55, and were differentiated for approximately 60 days at the time the experiments were conducted. hiPSCs were purchased from Coriell Institute (GM25256). Briefly, cells were cultured at 37 °C with 5% CO2. The monolayers of hiPSCs were differentiated into hiPSC-CMs by modulating Wnt signaling according to a small molecule-based cardiac differentiation strategy followed by metabolic lactate selection. Differentiated cells were reseeded in 8-well Ibidi µ-slides coated with Geltrex. Imaging was conducted under transmitted light illumination in an inverted Leica DMIRE2 microscope, within a OKOLab sample incubator chamber providing 37 °C, 5% CO2, 85% humidity. Images were acquired using an Andor iXon EMCCD camera operated at 100 Hz, yielding an effective frame time of approximately 26 ms. Kymographs to determine the beating rate were generated using imageJ.
Cell area measurements with confocal microscopy
To measure the effect of hypotonic treatment on cell area murine cardiomyocytes were seeded in 8-well Ibidi µ-slides with a density of 1,000 cells per well and labeled with Cell Mask™ Deep Red according to manufacturer’s protocol. XY and XZ movies of cells were then acquired on a confocal laser scanning microscope, Leica SP8, with a white-light laser at the wavelength of 633 nm and laser power of 5%. All measurements were conducted with an HC PLAP CS2 40×1.3 numerical aperture (NA) oil immersion objective (Leica). Movies were acquired at 30 seconds per frame with a hybrid detector in the range of 643 to 693 nm. Medium change to induce swelling was performed as described in previous sections. ImageJ was used to conduct thresholding and object detection on the cells and the extracted area was plotted with GraphPad Prism v. 9.5.1.
Linescan measurements with confocal microscopy
HEK293AD cells were seeded in 8-well Ibidi® µ-slides with a density of 25’000 cells per well and transfected with 0.25 µg Gαs-eYFP using the JetPrime® transfection kit according to manufacturer’s protocol. Linescans were conducted in a SP8 confocal microscope (Leica Microsystems) and analyzed as previously described using IgorPro 9 (wavemetrics)56.
FRET microscopy
HEK293 cells stably expressing Epac-SH187 sensor were seeded in 8-well Ibidi µ-slides with a density of 25’000 cells per well. Cells were washed twice in MES300 and imaged at room temperature. An inverted microscope (DMi8, Leica Microsystems), equipped with an x63 HC PL APO, 1.40-0.60 numerical aperture (NA) objective (Leica Microsystems), dichroic beamsplitter T505lpxr (Visitron Systems), xenon lamp coupled with a continuously tunable Visichrome high-speed polychromator (Visitron Systems) and a metal-oxide-semiconductor camera (Prime95B, Teledyne Photometrics) with a dual image splitter (OptoSplit II, Cairn Research), was used. Excitation wavelength of 445 nm was used, and fluorescence emission was simultaneously recorded at 470/24 nm and 535/30 nm. The movies were obtained at 5 seconds per frame for the number of frames needed for the cAMP concentrations to equilibrate. ImageJ was used to extract fluorescence intensity values from single cells, which were corrected for background and used to calculate FRET/CFP ratio.
Plate reader cAMP FRET measurements
HEK293 cells stably expressing Epac-SH187 sensor were seeded in black 96-well plates (Brand) with a density of 50’000 cells per well. Gαs-KO cells were transfected with either 2.5 µg Epac-SH187 or with 1.25 µg Epac-SH187 plus 1.25 µg Gαs 24 h prior to seeding in 96-well plates. 24 h after seeding cells were washed twice with 90 µl MES300 per well. After 5 min incubation at 37 °C, baseline measurement was conducted in a Neo2 plate reader (Biotek) using 420 nm excitation and 485 nm / 540 nm emission filters. As the second step, 60 µl MES300 were removed from each well and replaced with 60 µl of either MES300 or ES150 for the “control” and “swelling” conditions respectively and the plate was measured for 15 minutes. Afterwards the desired dilution series with increasing concentrations of ligand was added (10 µl per well of a 10x concentration in MES300) and the plate was measured again for 15 minutes. At last, a mix of forskolin/IBMX in MES300 was added to an end concentration of 10 µM forskolin and 100 µM IBMX and measured for 10 more minutes. The change in acceptor/donor ratio was normalized to 0% baseline and 100% forskolin/IBMX and plotted in GraphPad Prism v.9.5.1. For the concentration response curves “Dose-response stimulation fit (three parameters)” was applied.
TIRF microscopy
HEK293AD cells were seeded in 8-well Ibidi® µ-slides with a density of 25’000 cells per well and transfected with JetPrime® transfection according to manufacturer’s protocol. Following cDNA amounts per well were used: for Nb80 recruitment – 0.225 µg Snap-β2AR and 0.025 µg Nb80-eYFP; for Nb37 recruitment – 0.05 µg Snap-β2AR, 0.15 µg Gβ1−2A-Gγ2-IRES-Gαs and 0.05 µg Nb37-eYFP; Transfected cells were labeled with 1 µM SNAP-Surface 647 dye (NEB) for 30 min followed by washing two times for 10 min with 300 µl MES300 per well. After labeling, cells were subsequently taken for imaging to an Attofluor cell chamber (Fisher Scientific) in MES300. A TIRF illuminated Eclipse Ti2 microscope (Nikon), equipped with a x100, 1.49 NA automated correction collar objective and 405-, 488-, 561-, 647-nm laser diodes coupled via an automated N-Storm module and four iXon Ultra 897 EMCCD cameras (Andor), was used. Objective and cell chamber were kept at 37 °C during imaging. The automated objective collar was on, and hardware autofocus was activated. Movies were acquired at 4 s per frame for 400 frames. After a baseline measurement of 50 frames increasing concentrations of isoproterenol were added to the imaged well in 50 frame steps. Prior to each isoproterenol addition 30 µl of solution was removed from the well and 30 µl of 10x isoproterenol in either MES300 or MES200 was applied. ImageJ was used to extract fluorescence intensity values, which were corrected for background and normalized to 0% baseline and 100% 10 µM isoproterenol stimulation. In case of F-actin content measurement HEK293AD cells were transfected with Lifeact-eGFP and imaged as described above at 30 seconds per frame.
Single-molecule TIRF microscopy
HEK293AD cells were seeded on glass coverslips in 6-well plates at a density of 250,000 cells per well. To ensure low levels of receptor expression, necessary for single-particle imaging, transfection with 2 µg Snap-β2AR was performed with the JetPrime® transfection kit 3-4 hours before imaging. SNAP-tagged receptors were labeled with 1 μM SNAP-surface 549® dye and washed twice for 15 minutes with the imaging buffer (MES300). Imaging was done using AttoFluor chambers with the objective and the sample kept at 20 °C. The movies of receptor diffusion were then recorded via the Cy3 emission channel under illumination by the 561 nm laser at 100% laser power. Exposure time was set to 30 ms and movie length limited to 1000 frames to avoid excessive photobleaching.
Single particle tracking analysis
Obtained TIRF movies were analyzed by uTrack software57, loaded to the MATLAB environment. Point source detection and tracking analysis (incl. segment merging/splitting; maximum gap to close – 1 frame) modules were executed and resulting files, containing movie trajectories were loaded to MATLAB. Trajectories were filtered by length (between 3 and 100 frames) and by a manually defined ROI, selecting basolateral membranes of the cells. Mean square displacements for each movie were calculated over a time lag of 100 frames. A linear regression was fit to the first five frames of every resulting MSD movie and the slope was divided by four to obtain the diffusion coefficient.
Plate reader ligand binding assays
HEK293 cells were seeded in 10 m culture plates, co-transfected with 0.5 µg Snap-β2AR, 0.5 µg Nb37-eYFP and 1.5 µg Gβ1−2A-Gγ2-IRES-Gαs-CFP after 24 h and reseeded in black 96-well plates with a density of 50,000 cells per well another 24 h later. The plate was incubated with 100ul MES300 containing 2 nM JE1319. As the second step, 60 µl MES300 was removed from each well and replaced with 60 µl of either MES300 or ES150 (both containing 2 nM JE1319) for the “control” and “swelling” conditions respectively and the plate was incubated for further 15 minutes. Afterwards the desired dilution series with increasing concentrations of isoproterenol was added (10 µl per well of a 10x concentration in MES300) and the plate was incubated for 90 minutes and measured in a Neo2 plate reader (Biotek) using DualFP polarization filter and an 620 nm/680 nm excitation/emission filter. A G-factor of 0.12 was measured for the instrument. Fluorescence anisotropy was calculated as \(r=\left({I}_{\parallel }-{I}_{\perp }\right)/\left({I}_{\parallel }+2{I}_{\perp }\right)\) and plotted in GraphPad Prism v.9.5.1. For the concentration response curves “Dose-response inhibition fit (three parameters) was applied with min and max being constrained to 0% and 100% respectively.
Plate reader Gi-FRET sensor assay
HEK293 cells were seeded in 10 cm cell culture dishes at a density of 4 × 106. Cells were co-transfected with 0.5 µg SNAP-µOR and 2 µg Gi2 FRET sensor 24 h after seeding, as described in previous sections; 24 h after transfection, cells were trypsinized and transferred into black 96-well plates (Brand), at a density of 50,000 cells per well; 16 h to 24 h later, cells were washed with MES300, and then 90 μl of MES300 was added to each well. After 10 min incubation at 37 °C, measurement was performed at 37 °C using a Synergy Neo2 Plate Reader (Biotek) using a CFP/YFP FRET filter set. After basal FRET measurement, medium was changed as described above and after 15 minutes of further measurement 10 µl of ligand solution were applied to each well. The change in acceptor/donor ratio was normalized to 0% baseline and plotted in GraphPad Prism v.9.5.1. For the concentration response curves “Dose-response stimulation fit (three parameters)” was applied.
AlphaScreen cAMP detection assay
HEK293 cells were seeded in transparent 96-well plates (TPP) at a density of 25,000 cells per well. After 24 h the growing medium was exchanged to 100 µ of either MES300 or MES200 and increasing concentrations of isoproterenol were added to corresponding wells. After 10 minutes of stimulation, the plate was frozen at −80 °C for 15 minutes, resulting in cell lysis. The contents of the plate were then thawed at room temperature and 10 µl from each well were respectively transferred to a 384-well white OptiPlate™ plate (Revvity). cAMP standard solutions, acceptor- and donor-bead solutions were prepared and added to the plate according to the manufacturer’s protocol. The readouts of fluorescence intensity were performed at a VICTOR Nivo™ plate reader (Revvity) according to a measurement protocol provided by the manufacturer. The resulting cAMP concentrations were calculated and plotted in GraphPad Prism v.10.2.1. For the concentration response curves “Dose-response stimulation fit (three parameters)” was applied.
Statistical analysis
Statistical analysis was performed in GraphPad Prism v.10.2.1, statistical significance was defined as P < 0.05 and the significance values are indicated in the following P value style: 0.1234 (ns), 0.0332 (*), 0.0021(**), 0.0002 (***), <0.0001 (****).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.