Datasets
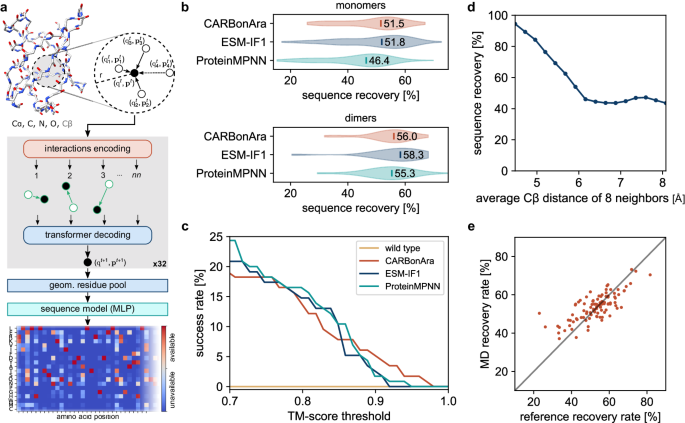
The training dataset is composed of ~370,000 subunits, the validation dataset of ~100,000, all downloaded from RCSB PDB, labeled as the first biological assembly (95% of which are annotated as such by the authors or automatically predicted as such and then confirmed by the authors). The test dataset is composed of ~70,000 subunits (single-chain proteins) with no shared CATH domains with the training set and less than 30% sequence identity with the test set. Within the test dataset, we extracted subunits without any shared CATH domains and maximum 30% sequence identity with any training set of PeSTo ( ~ 370,000 subunits), ProteinMPNN ( ~ 540,000 subunits), or ESM-IF1 ( ~ 18,000 subunits). This comparison dataset is composed of 228 subunits: 76 monomers, 37 dimers, and other 22 multimers. Note that ProteinMPNN and ESM-IF1 both use CATH classification and 40% sequence identity clustering for training and testing.
Features and labels
During the processing phase, we kept only the backbone of proteins (Cα, C, N, O), disregarding the hydrogen atoms, while adding the virtual Cβ using the ideal angle and bond length in the same way as in ProteinMPNN8. The structures we used to train the model can contain any type of molecule, including water, ions, nucleic acids, and any other non-protein molecules. The input scalar state contains the one-hot encoded 30 most frequent atomic elements in the PDB database. The last one-hot channel represents any other or unknown element. The input vector state is initialized randomly drawn from an isotropic normal distribution. We incorporated the geometric features using the pair-wise distance matrices and normalized displacement vector tensor. The output of the model is prediction confidence for each amino acid position among the 20 possible amino acid types (Supplementary Fig. 11). These types are represented as one-hot encoded labels. We optimized the model for multi-class classification of the 20 possible amino acids per position using a binary cross-entropy loss function.
Protein structure transformer architecture
The deep learning architecture of CARBonAra is almost identical to PeSTo21. We first embedded the input features into an input state size (S) of 32 using a three-layer neural network with a hidden layer size of 32. We then applied sequentially four sets of eight geometric transformers (S = 32, Nkey = 3, Nhead = 2), see Supplementary Algorithm 1 and Supplementary Fig. 1. The four sets of eight geometric transformers have a corresponding increasing number of nearest neighbors (nn = 8, 16, 32, 64). In instances where the number of atoms is less than the set number of nearest neighbors (nn), we assigned any additional non-existent interactions to a sink node. We configured this sink node with a constant scalar and vector state of zero. Next, the geometric residue pooling module reduced the atomic-level encoding of the structure into a residue-level description. This aggregation used a local multi-head mask on the atoms that constitute each residue (S = 64, Nhead = 4). Finally, we employed a multi-layer perceptron in the last module, which used three layers of hidden size (S = 64) to decode the state of all residues and computed the prediction, consequently generating a confidence score of the 20 possible amino acids through a sigmoid function ranging from 0 to 1.
Training
We trained our neural network architecture for 16 days on a single NVIDIA V100 (32 GB) GPU. To manage memory usage during training, we limited the subunits to a maximum of 8192 atoms (approximately 100 kDa), excluding hydrogen atoms. Furthermore, subunits containing fewer than 48 amino acids were not considered in the training process. The post-processing effective dataset contains 86610 structures in the training dataset and 24601 structures in the validation dataset.
Sequence sampling
We sampled the optimal sequence by taking the highest confidence amino acid per position from the prediction. To generate sequences with minimum sequence identity to the scaffold, we selected the highest confidence predicted possible amino acid above the positive prediction threshold of 0.5, which is not the original amino acid from the scaffold. The original amino acid is only used in the sequence generated if it is the only possible option within the positive predictions. Our criterion for defining the similarity between two amino acids relies on their BLOSUM37 62 score. We considered them as similar if this score is above zero. We sampled low sequence similarity to the original scaffold by restricting the positively predicted amino acids. When the options were available, we selected the amino acid with the highest BLOSUM 62 score below or equal to zero compared to the reference scaffold. If there are no options with a BLOSUM 62 score below or equal to zero, we sampled the positive predicted amino acid with the lowest BLOSUM 62 score. We noticed that taking only the minimum BLOSUM 62 similarity score generates sequences with a bias towards special amino acids (i.e., cysteine, proline, glycine). We performed a BLAST26,38 analysis to measure the novelty of the generated sequences with minimum identity and low similarity using the non-redundant protein sequences database with an expected value (E-value) cut-off at 100.
Alphafold and alphafold-multimer validation
In the case of the monomers, we sampled the highest confidence sequence from the predictions of CARBonAra for 142 subunits of the testing dataset. We also generated sequences using ProteinMPNN and ESM-IF1, both with a sampling temperature of 1e–6. We modeled the structures from the generated sequences with ColabFold39 (version 1.5.2) using the alphafold2_ptm model, in single-sequence mode and with 3 recycles40. In the case of the dimers, we generated sequences for one subunit, given the sequence of the other subunit. We sampled the sequence with the highest confidence from CARBonAra for the 31 dimers in the testing dataset for a total of 62 complexes with conditioning. We predicted the structures from the generated sequences with ColabFold (version 1.5.2) using the alphafold2_multimer_v2 model, in single-sequence mode and with 5 recycles41. To evaluate the sampling flexibility of CARBonAra, we sampled sequences with maximum identity, minimum identity, and low similarity using CARBonAra’s multi-class predictions. In this case, we used AlphaFold using multiple sequence alignment since a low sequence identity or similarity negates the sequences matching the reference scaffold in the multiple sequence alignment information. We assessed the predicted structures from the generated sequence with the original scaffold using the TM-score42 and Local Distance Difference Test43 (lDDT) on the Cα coordinates.
Molecular dynamics simulations
We selected 20 complexes from the Protein-Protein Docking Benchmark 5.0 dataset44 based on structure resolution and parameterization difficulty. For each complex, we conducted a standard 1 µs-long molecular dynamics (MD) simulation in the NPT ensemble (at 1 atm and 300 K, following a 2 ns NVT equilibration and using settings as per ref. 45) for the bound receptor, unbound receptor, bound ligand, and unbound ligand. We set up all systems using Amber ff14SB46 and its recommended TIP3P water model, running MD simulations with Amber1647. For the 80 (single chain structure) MD, we sampled 500 frames for each simulation and computed the average prediction confidence.
Comparison with deep sequencing
As a case study, we showed that CARBonAra’s residue-wise estimated probabilities (Supplementary Fig. 11) can be reliably correlated with experimentally determined mutations for the class A β-lactamase TEM-1. This widely studied enzyme has been subjected to deep mutagenesis by Deng et al., where the authors analyzed the effect of consecutive triple point mutations along the whole extension of the protein, covering all 20 naturally occurring amino acids per position33. The generated libraries were introduced in E. coli and selected based on ampicillin resistance. These data were used to compute a statistical change in free energy of binding (ΔΔGstat) of mutation of all wild-type residues in the protein. This value was calculated as ΔΔGstat = RT ln(pwt/pmut), where pwt and pmut are the probabilities of finding the wild-type and mutant amino acids, respectively, at the analyzed sequence position. Deng et al. also performed the same calculation on a MSA of 156 sequences of class A β-lactamases, to compare the conservation profile of this family with the requirements imposed by the mutagenesis assays. Aiming at assessing CARBonAra’s ability to recover evolutionary-related residue profiles, we used its residue-wise estimated probabilities to compute the ΔΔGstat per position of TEM-1. We used two structures of TEM-1 as input for the model: TEM-1 in the apo state (PDB ID: 1JTG, removing all non-protein atoms) and TEM-1 retaining its catalytic water and β-lactam nitrocefin at the catalytic pocket. Docking of this ligand to TEM-1 was carried out with AutoDock Vina48 and the analyzed pose was selected based on the proximity of the carbonyl group of the β-lactam ring to the catalytic residue S70. We then calculated Pearson’s correlation coefficient (ρ) of the deep sequencing and CARBonAra’s estimated ΔΔGstat per sequence position.
Nitrocefin docking to TEM-1
This step was necessary because there are no structures currently available of nitrocefin complexed with TEM-1. The docking was carried out with AutoDock Vina48. We obtained the 3D coordinates of nitrocefin from the PubChem database (PubChem CID: 6436140) and used a search space of size 40x40x40 Å centered on the enzyme’s active site (determined by visual inspection). The exhaustiveness parameter was set to 200 and 30 models were generated. The analyzed pose was selected based on the proximity of the carbonyl group of the β-lactam ring to the catalytic residue S70. We also looked for interactions between nitrocefin and residues R244 and N132, known for the stabilization of cephalosporins in TEM-149,50.
Sequence sampling using imprinting
CARBonAra produces matrices (i.e., position-specific scoring matrices, Fig. 1a) that score preferences for each of the 20 amino acids at each position of the designed sequence. The prediction confidence can be converted into a probability (Supplementary Fig. 11). Imprinting in CARBonAra allows the specification of arbitrary sequence information to any position in the backbone scaffold as prior information for the prediction. To efficiently sample the sequence space, we developed an imprinting protocol where first we predict and select the amino acids with the highest confidence score at each sequence position. Afterward, we randomly select between 10 to 90% of these amino acids to imprint on their corresponding backbone positions. We then use the backbone with imprinted information to run a second CARBonAra prediction, from which we select the amino acids with the highest confidence score at each position to build the designed sequence. This strategy promotes sequence diversity while providing high-confidence amino acids per position by scrambling each time the set of amino acids is imprinted after the first prediction. When designing TEM-like enzymes, we used as the input its structure constrained by the presence of the catalytic water and nitrocefin. We predicted a total of 900 sequences within the imprinting range 10-90%, and first filtered for sequences that were able to recover TEM-1’s catalytic triad plus two residues known to accommodate β-lactams in the active site (namely S70, K73, N132, E166, and R244). These sequences were then modeled with AlphaFold in single-sequence mode and ranked based on the highest plDDT. Only the first 10 top-ranked sequences were selected for in vitro validation. The sequences of the 4 that were soluble underwent further experimental validation and are reported in Supplementary Dataset 1.
Material
We purchased HEPES from Chemie Brunschwig AG (Basel, CH), isopropyl β-d-1-thiogalactopyranoside (IPTG) from Huberlab (Aesch, CH) and all other chemicals from Merck (Darmstadt, DE), unless specified.
Protein expression and purification of TEM-like designs
The coding sequences for WT TEM-1 and all the variants were optimized for Escherichia coli (E. coli) expression and cloned into the pET28a(+)-TEV expression vector (Genscript) between NdeI and XhoI, such that the resulting constructs have a N-terminal His-tag followed by a TEV cleavage site. The plasmids were transformed in Rosetta (DE3) cells (Promega). Protein expression was induced by the addition of 1 mM IPTG when the cells reached an optical density of 0.6 and subsequent growth overnight at 20 °C. Cell pellets were resuspended in lysis buffer (20 mM HEPES, pH 7.5, 500 mM NaCl and cOmplete™ Protease Inhibitor Cocktail (Roche)), lysed using sonication, and centrifuged (20000xg for 35 min at 4 °C). The supernatant was applied to a HisTrap HP column (Cytiva) previously equilibrated with lysis buffer. The proteins were eluted with a continuous gradient over 40 column volumes of elution buffer (20 mM HEPES, pH 7.5, 500 mM NaCl, 500 mM Imidazole). Subsequently, pure fractions were additionally purified by Size Exclusion Chromatography (Superdex S200 Increase, Cytiva) in 20 mM HEPES, pH 7.5, 300 mM NaC, 1 mM TCEP. The proteins were flash-frozen in liquid nitrogen and stored at –20 °C.
Size exclusion chromatography coupled to multi-angle light scattering
The molecular weights of the constructs were determined by size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS). The mass measurements were performed on a Dionex UltiMate3000 HPLC system equipped with a 3 angles miniDAWN TREOS static light scattering detector (Wyatt Technology). The sample volumes of 5−10 μl at a concentration of 8 mg/mL, were applied to a Superose 6 Increase 3.2/300 column (Cytiva) previously equilibrated with 20 mM HEPES pH 7.5, 300 mM NaCl at a flow rate of 0.08 mL/min. The data were analyzed using the ASTRA 6.1 software package (Wyatt technology), using the absorbance at 280 nm and the theoretical extinction coefficient for concentration measurements.
Circular dichroism
CD spectra were collected on 10 μM protein solutions in 50 mM Tris pH 7.5, 150 mM NaCl, using a Chirascan CD polarimeter (Applied Photophysics, UK) in 1 mm path cuvettes, at 20 °C. Thermal denaturation curves were acquired by heating the sample from 20 to 98 °C every 1 °C, collecting full spectra and plotting the trace at 222 nm.
NMR spectroscopy
The 1H, 15N HSQC spectrum of TEM design D4 was obtained on a 15N–labeled 250 µM sample prepared in MES pH 6.5 with 200 mM NaCl and 10% 2H2O, at 318 K. It was acquired in a Bruker Avance II 800 MHz (1H frequency) spectrometer equipped with a CPTCIXYZ cryoprobe, using a standard 15N HSQC pulse program with water suppression and sensitivity enhancement, 256 increments in the indirect dimension, and a recycle delay of 1 s.
β-Lactamase activity
TEM−1 and 4 TEM-like designed proteins were incubated at the indicated concentrations (Fig. 2f) with 200 µM nitrocefin in 20 mM HEPES, pH 7.5, 300 mM NaCl at either 30 or 70 °C in a 1 mm path length cuvette while monitoring the absorbance at 485 nm changing over time using a Chirascan CD polarimeter (Applied Photophysics, UK). All temperatures reported involve thermal equilibration of cuvette, buffer, nitrocefin and protein at the indicated temperature for 5 min before mixing.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.