Chemical probes and inhibitors
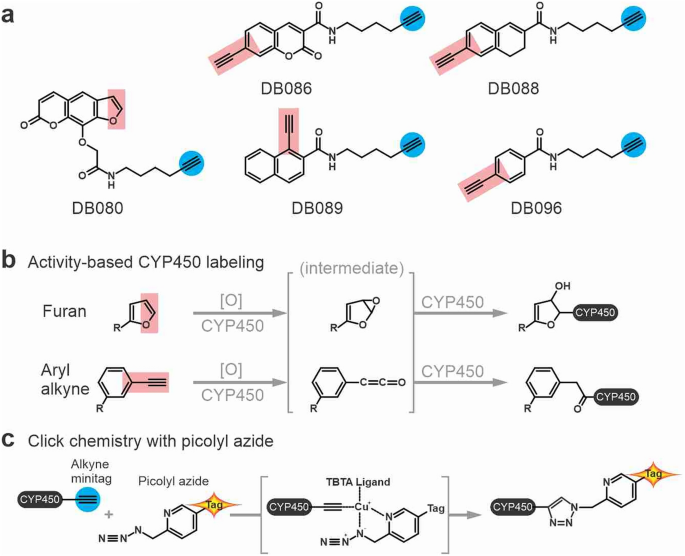
The synthesis of probes DB088, DB089 and DB096 was carried out with minor alterations to the established routes as described previously11. Starting from the parent acids, a sequence involving (i) protection as the respective methyl esters; (ii) Sonogashira reactions to install the TMS-protected acetylene; and (iii) global deprotection, resulted in the alkynyl acids. These acids underwent smooth conversion to DB088, DB089, and DB096 using amide coupling procedures.
The synthesis of DB086 retained the general strategy for probe synthesis with an additional step required to build the coumarin core and activate the phenolic position. In order to build the coumarin core, the Knoevenagel condensation of 2,4-hydroxybenzaldehyde with diethyl malonate was employed. From this, triflation of the phenolic position enabled the established Sonogashira reaction, deprotection and amide coupling to be carried out giving DB086.
Similarly, DB080 could be produced with only minor alterations to established conditions. Demethylation of methoxsalen was carried out with BBr3, the resulting phenol underwent smooth conversion to the requisite ester upon reaction with methyl bromoacetate. Deprotection gave access to the DB080 precursor which gave DB080 after amide coupling.
A de novo approach to DB143 was employed on account of stability issues faced during the Krapcho decarboxylation employed previously11. Propiolic acid was hydrobrominated to produce the respective (E)-vinylbromide, this was subsequently reduced with LiAlH4 to give the respective alcohol. This was amenable to a Sonogashira reaction with TIPS-acetylene to give the TIPS-protected enyne that was able to undergo conversion to DB143 using a route analogous to that published earlier11, requiring only an additional deprotection step to remove the TIPS-protecting group.
For further details on probe synthesis, see Supplemental Methods. Aliquots of the probes are available upon request. The P450 inhibitor 1-ABT (A3940-50MG) and agrochemical benoxacor (46001-50MG) were purchased from Sigma-Aldrich. NADPH was purchased from MELFORD (N20140-0.1).
Molecular cloning
Plasmids and Primers are summarized in Supplementary Tables S1 and S2, respectively. Golden Gate Modular Cloning kit33 and Golden Gate Modular Cloning Toolbox for plants34 were used for cloning. The sequence encoding mouse MmCyp1a2 and maize ZmCYP81A9 were codon-optimised for in planta expression and synthesized as a GeneString (GeneArt, ThermoFisher Scientific, Supplementary Table S1). Using BpiI restriction sites, these ORFs were first cloned into the level 0 module GG1-01, and transferred into pJK268c35 together with GG1-55, GG1-57, GG1-70 or GG1-78 in a BsaI ligation reaction. Generated plasmids were amplified by transformation into E. coli and the inserts were confirmed by sequencing.
The sequences encoding Z. tritici P450s (CYP5078B1, CYP5080H1, CYP52R1 and CYP539A6) were amplified from Z. tritici complementary DNA (cDNA) using the forward and reverse primer pairs summarised in Supplementary Table S2. Using BsaI restriction sites, PCR products were combined with GG1-55, GG1-57, GG1-70 or GG1-7834 to assemble the corresponding expression plasmids. Generated expression plasmids were transformed into E. coli for their amplification and sequencing.
Binary plasmids were transformed into Agrobacterium tumefaciens strain GV3101, carrying the pMP90 helper plasmid36. Agrobacterium transformants were selected on LB plates (10 g/L NaCl, 10 g/L Tryptone, 5 g/L yeast extract, 15 g/L agar) containing 50 μM gentamycin, 25 μM rifampicin and 50 μM kanamycin. Single colonies were picked and grown in liquid LB (10 g/L NaCl, 10 g/L Tryptone, 5 g/L yeast extract) containing the same antibiotic used before. Glycerol stocks of the selected transformants were prepared mixing Agrobacterium cultures with 50% sterile glycerol in a 1:1 ratio.
Plant growth and agroinfiltation
Nicotiana benthamiana plants (LAB accession, collected by Cleland in 193637, specimen AD957110022 at State Herbarium of South Australia in Adeleide) were grown in the greenhouse at the University of Oxford on soil at 21 °C under long day conditions (16/8 h light/dark regime) and were used for agroinfiltrations when 4â5 weeks old. Use of N. benthamiana is compliant with the Nagoya protocol because this species was collected in 1939. Growing N. benthamiana is compliant with institutional, national and international guidelines and legislation since N. benthamiana is neither an endangered species nor at risk of extinction according to IUCN. Agrobacterium cultures (GV3101 pMP90) were grown overnight (approximately 18 h) at 28 °C with agitation in LB media containing 50 μM gentamycin, 25 μM rifampicin and 50 μM kanamycin. Bacteria were collected by centrifugation at 1000Ãg for 10 min at room temperature (RT), washed with infiltration buffer (10 mM MES, 10 mM MgCl2, pH 5.6 and 100 μM acetosyringone) and diluted to OD600â=â0.5. Agrobacterium were incubated at 28 °C for 2 h and hand-infiltrated into two fully-expanded leaves using a needle-less 10 ml syringe.
ZmCYP81A9 expression in yeast
The ORF of CYP81A9 was codon-optimized for expression in Saccharomyces cerevisiae and synthesized and cloned into pYES3 vector by TWIST Bioscience (CA, USA) and transformed into the WAT11 strain18. For yeast cultivation, yeast was grown in selective agar media containing adenine {6.7 g/L yeast nitrogen base without aminoacids (Sigma Y0626), 0.64 g/L complete supplement mixture minus tryptophan (CMS-Trp), 20 g/L glucose (Sigma G8270), 20 g/L bacto agar (BD Difco, 214050), 0.04 g/L adenine sulphate (Sigma A-9126)} at 30 °C for 48 h. A single yeast colony was inoculated in 25 ml of selective liquid media {6.7 g/L yeast nitrogen base without aminoacids (Sigma Y0626), 0.64 g/L complete supplement mixture minus tryptophan (CMS-Trp), 20 g/L glucose (Sigma G8270), 0.04 g/L adenine sulphate (Sigma A-9126)} and grown at 30 °C, 180 rpm, for 24 h. Next, 6 mL of the yeast culture was inoculated in 300 mL of YPGE containing adenine {10 g/L yeast extract (Sigma 92144), 10 g/L peptone (Melford P20P240), 30 mL/L ethanol, 20 g/L glucose and 0.04 g/L adenine sulphate (Sigma A-9126)} and incubated with agitation (180 rpm) at 30°C for 30 h. To induce protein expression, yeast cells were washed with water in sterile conditions, resuspended in 300 mL of YPLE containing adenine {10 g/L yeast extract (Sigma 92144), 10 g/L peptone (Melford P20P240), 30 mL/L ethanol, 20 g/L d-galactose, 0.04 g/L adenine sulphate (Sigma A-9126)} and incubated with agitation (180 rpm) at 30 °C for 16 h. Yeast cells were harvest by centrifugation and used for microsome isolation.
Yeast microsomes were prepared by washing the yeast cells in TEK buffer (50 mM TrisâHCL pH 7.5, 1 mM EDTA, 100 mM KCl). Yeast cells were resuspended in ice-cold TESBâ+âDTT buffer (50 mM TrisâHCL pH 7.5, 1 mM EDTA, 600 mM d-Sorbitol, 30 mM DTT) and incubated for 10 min at room temperature. Afterwards, yeast cells were centrifuged at 5000Ãg, resuspended in ice-cold TESB (50 mM TrisâHCL pH 7.5, 600 mM d-Sorbitol) and ground using 0.5 mm glass beads and a bead mill homogenizer (BioSpec BeadBeater 1107900-105). The cell extract was filtered through a nylon cloth and centrifuged at 16,000Ãg for 10 min. The supernatant was ultra-centrifuged at 100,000Ãg for 1h. Pellet containing the microsomal membranes was resuspended in TEG buffer (50 mM TrisâHCL pH 7.5, 1 mM EDTA, 20% glycerol) and protein concentration was adjusted to 2 mg/ml by Bradford protein assay. Microsomes were kept atâââ80 °C until used.
Protein extraction and microsomal fraction isolation from plant tissues
Agroinfiltrated N. benthamiana leaf tissue was collected at 4 days post-infiltration (dpi). Leaf tissue was frozen in liquid nitrogen and ground to fine powder with a mortar and pestle. Total proteins were extracted in cold homogenization buffer (100 mM potassium phosphate buffer pH 7.5, 400 mM sucrose, 1% (w/v) polyvinylpolypyrrolidone (PVPP) and 4 mM β-mercaptoethanol). The extracts were left for 20 min rotating at 4 °C for complete homogenization and centrifuged at 5000Ãg for 20 min at 4 °C. Supernatants containing soluble proteins were filtered through a layer of miracloth filter and ultra-centrifuged at 100,000Ãg for 1 h. Pellets containing the microsomal membranes were resuspended in resuspension buffer (100 mM potassium phosphate buffer pH 7.5 containing 20% glycerol). Total protein concentration was determined by DC protein assay Kit (Bio-Rad) with bovine serum albumin standards, protein concentration was adjusted to 0.75 mg/mL for each sample and the resulting samples were used for labelling without freeze thawing.
Activity-based protein profiling and click chemistry
All probes were prepared at 0.5â2 mM stock solutions in dimethyl sulfoxide (DMSO). Protein samples of 50 μL were incubated with or without the indicated probes in the presence or absence of 1 mM NADPH for 1 h at the indicated probe concentration (usually 5â10 μM). Labelling reactions were stopped by adding 1:4 (v/v) cold acetone and precipitating by 3 min centrifugation at maximum speed using a benchtop centrifuge. Protein pellets were resuspended in 44.5 μL of PBS-SDS buffer {phosphate-buffered saline (PBS) pH 7.4 with 1% (w/v) sodium dodecyl sulfate (SDS)} and heated for 5 min at 90 °C. For click chemistry, 44.5 μL of labelled proteins were treated with 2 μM Cy5 Picolyl Azide (Click Chemistry Tools, 50 μM stock in DMSO) or Fluorescein Picolyl Azide (Click Chemistry Tools, 50 μM stock in DMSO) and a premixture of 100 μM tris((1-benzyl-1H-1,2,3-triazol-4-yl) methyl)amine (TBTA; 3.4 mM stock in DMSO:t-butanol 1:4), 2 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP; 100 mM stock in water) and 1 mM copper(II) sulfate (CuSO4; 50 mM stock in water). Samples were incubated for 1 h at RT in the dark and quenched by acetone precipitation. Protein pellets were resuspended in 50 μL of 2Ãâgel loading buffer (100 mM TrisâHCl pH 6.8, 200 mM dithiotreitol, 4% SDS, 20% glycerol and 0.02% bromophenol blue) and heated for 5 min at 90 °C.
ZmCYP81A9 activity assay with bentazon
Microsomes of yeast expressing CYP81A9 (17 µL of 2 mg/mL) were incubated with 200 μM bentazon in the presence or absence of 1 mM NADPH and in a final volume of 100 μL of 50 mM TrisâHCl pH 7.5 buffer, at 28 °C for 2 h. For inhibition experiments, yeast microsomes were pre-incubated with 300 μM P450 probes or 1-ABT for 10 min in the presence of 1 mM NADPH before adding 200 μM bentazon. Reactions were stopped by adding 100 μL acetonitrile: HCl (99:1) and proteins were precipitated by 3 min centrifugation at maximum speed using a benchtop centrifuge. Supernatants were diluted by adding 1:5 (v/v) acetonitrile in glass vials. Samples were analysed by LCâMS using the electrospray ionization interface (ESI) in negative mode. Bentazon and hydroxybentazon were detected at retention times of 1.57 and 1.49 min, respectively. Data is expressed in amount of hydroxylated bentazon detected (Peak area, AU) in the incubated microsomes.
In-gel fluorescence scanning and western blot
Labelled proteins were loaded on 10â12% SDS-PAGE gels and separated at 150 V. Fluorescence scanning of acrylamide gels was performed on an Amersham Typhoon 5 scanner (GE Healthcare Life Sciences) with Cy2 and Cy5 settings using the 488nm and 635nm lasers, respectively. Scanned gels were stained with Coomassie Brilliant Blue G-250. Image J was used for the quantification of the signals.
For western blot analysis, proteins separated in SDS-PAGE gels were transferred onto a PVDF membrane using Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were blocked in 5% (w/v) milk in TBS-T (50 mM TrisâHCl pH 7.6, 150 mM NaCl and 0.1% Tween-20) overnight at 4 °C, incubated in 1:5000 anti-GFP-HRP (Abcam, ab6663) for 1.5 h at RT and washed in TBS-T for 5 times for 5 min and in TBS for the last wash. Clarity Western ECL substrate (Bio-Rad) was used for chemiluminescent protein detection. When performing streptavidin-HRP protein blots, samples were processed the same way except that membranes were blocked in 1% (w/v) BSA in TBS-T and incubated in 1:1000 streptavidin-HRP (Sigma, S2438).
Filter-aided sample preparation (FASP) for mass spectrometry
Mouse liver microsomes (purchased from Thermo: GIBCO Mouse (CD-1) Microsomes) were diluted to 1 mg/mL protein concentration. YM-10 microcon centrifugal filters (Millipore) were washed with 200 μL 0.1% trifluoroacetic acid (TFA) in 50% acetonitrile (ACN) and centrifuged at 15,000Ãg for 15 min at room temperature. Filters were pre-equilibrated with 200 μL of 8 M urea in 100 mM triethylammonium bicarbonate (TEAB) pH 8.5 and centrifuged at 15,000Ãg for 15 min at room temperature. Afterwards, an equivalent of 200 μg total protein were mixed with 200 μL of 8 M urea in 100 mM TEAB, pH 8.5 in the pre-treated filters. Filters were centrifuged at 15,000Ãg for 40 min at room temperature and retained proteins were washed with 200 μL of 8 M urea in 100 mM TEAB a total of 4 times. Protein reduction and alkylation were achieved by sequential incubation of the concentrates in 200 μL of 8 M urea in 100 mM TEAB containing tris(2-carboxyethyl) phosphine hydrochloride (TCEP; 10 mM final concentration, 30 min incubation at room temperature) and 2-chloroacetamide (50 mM final concentration, 30 min incubation at room temperature in the dark) followed by centrifugation. The concentrates were washed with 6 M urea in 50 mM TEAB a total of 2 times and subjected to proteolytic digestion with 2.5 μg of lysyl endopeptidase (Lys-C from FUJIFILM Wako, in 50 mM TrisâHCL pH 8, 4 h incubation at 37 °C) and 5 μg of trypsin (Trypsin Gold from Promega, in 50 mM TEAB pH 8.5, overnight incubation at 37 °C). Digested peptides were collected by centrifugation, and filters were washed with 150 μL 0.1% TFA and 150 μL 0.1% TFA in 50% ACN. The combined filtrates were dried by vacuum centrifugation and submitted for MS analysis.
Large-scale labelling, affinity purification
Microsomal fractions (1.5 mL of 0.75 mg/mL) were incubated with or without the indicated probes (20 μM) in the presence or absence of 1 mM NADPH for 1 h. Labelling reactions were stopped by adding 4 volumes of ice-cold methanol, 1 volume of ice-cold chloroform and 3 volumes of water and centrifugation for 40 min (4000Ãg, 4 °C). Protein pellets were resuspended in 1 mL of PBS-SDS buffer by bath sonication and heated at 90 °C for 10 min. Labelled proteins were biotinylated with 30 μM azide-PEG3-biotin (Sigma-Aldrich, 2 mM stock in DMSO) using click chemistry as described above. Samples were incubated for 1 h at room temperature in the dark, quenched by the addition of 10 mM EDTA and proteins were precipitated via the methanol/chloroform method. Protein pellets were resuspended in 1.5 mL of 1.2% (w/v) SDS dissolved in PBS by bath sonication and diluted by adding 7.5 ml of PBS. The resulting solution was incubated with 100 μL of pre-equilibrated Pierce High Capacity Streptavidin Agarose beads (Thermo Fisher Scientific) for 2 h at room temperature. Agarose beads containing the labelled proteins were collected by centrifugation at 1000Ãg for 5 min at room temperature. Agarose beads were washed successively three times with 10 mL of PBS-SDS buffer, three times with PBS and a final wash with 10 mL of water.
On-bead trypsin and Lys-C digestion
For on-bead digestion, the captured agarose beads containing the labelled proteins were treated with 500 μL 6 M urea dissolved in PBS and 10 mM TCEP for 15 min at 65 °C. A final concentration of 20 mM 2-chloroacetamide (400 mM stock in water) was added and left incubated for 30 min at 35 °C in the dark. Reaction was diluted by the addition of 950 μL of PBS and the supernatant was removed by centrifuging the beads at 500Ãg for 2 min. 100 μL of Lys-C digestion solution (Walko, 5 μg of Lys-C was reconstituted in 200 μL 1 M urea, 50 mM TrisâHCl buffer pH 8) was added to the beads and left incubating at 37 °C for 3 h. 100 μL of trypsin digestion solution (Promega, 5 μL of reconstituted trypsin was dissolved in 100 μL 50 mM TrisâHCl buffer pH 8) was added to the beads and left incubating at 37 °C overnight. The supernatant containing the digested peptides was dried by vacuum centrifugation and send for MS analysis.
LCâMS/MS and peptide identification using MaxQuant
MS samples were analysed as described as follows. The acidified tryptic peptides were desalted using C18 Tips (Thermo Fisher Scientific, 10 μL bed, 87782) and peptide samples were eluted with 0.1% formic acid and analysed on an Orbitrap Elite instrument (ThermoFisher Scientific)38 attached to an EASY-nLC 1000 liquid chromatography system (Thermo Fisher Scientific). Samples were separated on an analytical column based on a fused silica capillary with an integrated PicoFrit emitter (New Objective) with Reprosil-Pur 120 C18-AQ 1.9 μm resin. Peptides were next separated using 140 min gradient of solvent and analysed with Orbitrap analyser (using Fourier Transform MS) at a resolution of 60,000 with the internal lock-mass option switched on39.
RAW spectra were submitted to an Andromeda search in MaxQuant (version 1.5.3.30) using the default settings40,41. Label-free quantification and match between-runs were activated24. The MS/MS spectra data were searched against the UniProt Mus musculus reference database (55,412 entries). All searches included a contaminants database (as implemented in MaxQuant, 245 entries). Enzyme specificity was set to âTrypsin/Pâ and the MS/MS match tolerance was set toâ±â0.5 Da. The peptide spectrum match FDR and the protein FDR were set to 0.01 (based on target-decoy approach) and the minimum peptide length was seven amino acids. For protein quantification, unique and razor peptides were allowed. Modified peptides were allowed for quantification with a minimum score of 40. Relative quantification of proteins between different MS runs is based exclusively on the label-free quantifications as calculated by the software MaxQuant, using the âMaxLFQâ algorithm24. Filtering of results was done in Perseus version 1.6.13.042. Briefly, data was filtered against contaminants and only those proteins found in at least one group in all two/three replicates were considered. Further analysis and graphs were performed using the Qfeatures and ggplot R packages. Imputed values were generated using a missing not at random (MNAR) and missing at random (MAR) mixed imputation over the whole matrix, and the fold change and adjusted P values (adj. P Val) were calculated using the fdr method over the three biological replicates. The correct clustering of the biological replicates by categorical groups was evaluated using principal component analysis (PCA) plots.
Confocal microscopy
Fluorescence microscopy was carried out using a Zeiss LSM 880 Airyscan confocal microscope with a C-Apochromat 40Ã/1.2 W Korr FCS M27 water-immersive objective. GFP and RFP fluorescence were detected with sequential line scanning. GFP fluorescence was excited with 2% laser power at 488 nm and emission detected with a PMT detector at 493 to 579 nm; while RFP fluorescence was excited at 561 nm and emission detected with GaAsP detector at 582 to 754 nm. For z-stacks 16-bit images were captured at a slice interval of 1.31 μm. Colocalization of GFP and RFP signals was quantified with JaCoP2 plugin in Fiji software package in single images in selected regions of interest (ROIs), without saturation of fluorescence signals.