Experimental model and culturing
HEK293S-derived cells (T501) were grown in Freestyle 293 Expression Medium containing 5% tetracycline-free fetal bovine serum (FBS) in vented shaking flasks at 37â°C, 5% CO2 and 120ârpm (550âxâg). Culture was scaled up sequentially, by inoculating at 1.5 Ã106 cells/mL and subsequently splitting at a cell density of 3.0âÃâ106âcells/mL. Finally, a final volume of 2âL of cell culture at a cell density of 4.5âÃâ106âcells/mL was used for mitochondria isolation, as previously described85.
mS29-KO cells were derived from HEK293T (CRL-3216) and cultured in complete DMEM supplemented with 20% FBS at 37â°C and 5% CO2. HEK293T (CRL-3216) were cultured in high-glucose, sodium pyruvate, L-glutamine Dulbeccoâs modified Eagle medium (DMEM, Life Technologies) supplemented with 10% FBS, 100âµg/mL uridine, 3âmM sodium formate, and 1x MEM non-essential amino acids complete DMEM medium). Analysis of mycoplasma contamination was routinely performed.
For the labeling of cells with deuterated Uridine required for SILNAS analysis, TK6 cells deficient in uridine monophosphate synthase (UMPS-/-) were cultured at 37â°C in RPMI-1640 medium containing 10% horse serum, 2âmM L-glutamine, penicillin (50âU/ml), streptomycin (50âμg/ml) and 0.1âmM 5,6-D2-uridine.
Generation of mS29 edited cell lines and plasmid transfections
To generate a stable human mS29-KO cell line in HEK293T cells (ATCC: CRL-3216), we used a pool of CRISPR/Cas9-mediated knockout cells generated by the genome editing company, Synthego (Synthego Corporation, Redwood City, CA). HEK293T cells were first tested to be negative for mycoplasma. Three guide RNAs targeting exon 5 of the mS29 gene (DAP3-202 transcript ID ENST00000368336.10) were selected for high specificity and activity to create premature stop codons through frameshift mutations in the coding region via insertions and/or deletions (Indels). Targeting exon 5 ensured that all mS29 transcript variants would be affected. Based on additional off-target analysis, the three specific guide RNAs selected (g1, g2, and g3) were: g1 5â²-GUGAAGCUUGCCUGAUGGUA-3â² [AGG]-PAM, g2 5â²-UAUAGCUGGAUAAGCAAAAC-3â² [TGG]-PAM, and g3 5â²-AUGCUUUCUCCCCUCAAAUA-3â²â [AGG]-PAM. The pool of knockout cells was then generated by electroporation of ribonucleoproteins (RNPs) containing the Cas9 protein and synthetic chemically modified sgRNA (Synthego) into the cells using Synthegoâs optimized protocol. The editing efficiency was assessed upon recovery, 48âh post-electroporation. Genomic DNA is extracted from a portion of the cells, PCR-amplified, and sequenced using Sanger sequencing. The resulting chromatograms are processed using Synthego Inference of CRISPR Edits software (ice.synthego.com). The pooled mS29-KO was then plated into 96-well plates to screen for single clones in-house. Single clone candidates were screened by immunoblotting to determine the steady-state levels of mS29 and the mtDNA-encoded COX2 protein as a surrogate of mitochondrial protein synthesis capacity. Clones that had undetectable mS29 and attenuated COX2 levels were further analyzed by genotyping. For genotyping, the edited region of mS29 was amplified using oligonucleotides mS29 exon 5 seq (-164) Forward: 5â-GGATAGATTTTCAAACTCAGTACCA-3â and mS29 exon 5 seq (+166) Reverse: 5â-TCCTGACTTCAGGCGATACG-3â, and subsequently cloned into the TOPO-TA vector (Thermo Fisher) for sequencing.
To establish a stable cell line reconstituted with mS29, we obtained the WT mS29 gene as a Myc/DDK-tagged ORF from Origene (Cat# RC223182). Using restriction sites AsiSI and PmeI, WT mS29-Myc/DDK was cloned into a mammalian vector with hygromycin as the selection marker. The vector used was pCMV6, in which mS29 gene expression was placed under the control of an attenuated version of the human cytomegalovirus (CMV) enhancer/promoter (â5) in which a deletion in the promoter sequence eliminates most transcription factor binding sites86. Furthermore, to ensure functionality of the recombinant mS29, the Myc/DDK tag was removed via the Q5 site-directed mutagenesis kit (New England Biolabs) using the oligonucleotides mS29 âMyc-DDK Forward: 5â-TAAACGGCCGGCCGCGGT-3â and mS29 Îmyc-DDK Reverse: 5â-GAGGTAGGCACAGTGCCGCTC-3â. Two μg of the construct containing mS29 was transfected to the HEK293T mS29-KO cell line using 5âμL of EndoFectin⢠Max (GeneCopoeia) pre-incubated in Opti-MEM (ThermoFisher). 72âh post-transfection, the medium was supplemented with 200âμg/mL hygromycin B for three weeks. We use the same protocol to establish stable cell lines reconstituted with mutant variants of mS29.
Mutant variants of the mS29-containing construct were generated using the Q5 site-directed mutagenesis kit (New England Biolabs). Mutagenesis primers were designed using the NEBaseChanger tool. Oligonucleotides used for mutagenesis include: mS29 K134A Forward: 5â-GGGAACAGGAGCAACCCTAAGTC-3â; mS29 K134A Reverse: 5â-TTCTCTCCATACAGAAGATATC-3â; mS29 Y173A Forward: 5â-GCAGTCCAGCGCCAACAAACAGC-3â; mS29 Y173A Reverse: 5â-AGAAGATCCCGACAATTTTTC-3â; mS29 R177A Forward: 5â-CAACAAACAGGCCTTTGATCAACCTTTAGAG-3â; mS29 R177A Reverse: 5â-TAGCTGGACTGCAGAAGA-3â; mS29 K245A Forward: 5â-AATTGTGCTGGCAGAGCTAAAGAGGC-3â; mS29 K245A Reverse: 5â-CCAACTGCATCTGTGGCG-3â; mS29 H291A Forward: 5â-AGCACTTGTTGCCAACTTGAGGAAAATGATG-3â; mS29 H291A Reverse: 5â-AATTCCTCGGGGGCAATC-3â; mS29 K295A Forward: 5â-CAACTTGAGGGCAATGATGAAAAATGATTG-3â; mS29 K295A Reverse: 5â-TGAACAAGTGCTAATTCC-3â; mS29 Y208A Forward: 5â- TCAAGAGAAGGCTGTCTGGAATAAGAG -3â; mS29 Y208A Reverse: 5â- ACTTTTATCTGGTTCAGG -3â; mS29 V209P Forward: 5â- AGAGAAGTATCCCTGGAATAAGAGAG -3â; mS29 V209P Reverse: 5â- TGAACTTTTATCTGGTTCAG -3â; mS29 D238A Forward: 5â- GAACGCCACAGCTGCAGTTGGAA -3â; mS29 D238A Reverse: 5â- CTCACCCGTGTTATGCCC -3. All mutant sequences were validated by Sanger sequencing using oligonucleotides â5pCMV6 Forward: 5â-CCTCTTCGCTATTACGCCAG-3â, mS29 cDNA (308) Forward: 5â-AACCAGCCCTAGAACTTCTGC-3â, and mS29 cDNA (829) Forward: 5â-GAAGATAAAAGCCCGATTGC-3â.
Mitoribosome purification
Mitoribosome purification was carried out based on the previously developed protocol for rapid isolation85. HEK293S-derived cells were harvested from the 2âL culture when the cell density was 4.2âÃâ106âcells/mL by centrifugation at 1000âg for 7âmin, 4â°C. The pellet was washed and resuspended in 200âmL Phosphate Buffered Saline (PBS). The washed cells were pelleted at 1000âg for 10âmin at 4â°C. The resulting pellet was resuspended in 120âmL of MIB buffer (50âmM HEPES-KOH, pH 7.5, 10âmM KCl, 1.5âmM MgCl2, 1âmM EDTA, 1âmM EGTA, 1âmM dithiothreitol, complete EDTA-free protease inhibitor cocktail (Roche) and allowed to swell in the buffer for 15âmin in the cold room by gentle stirring. About 45âmL of SM4 buffer (840âmM mannitol, 280âmM sucrose, 50âmM HEPES-KOH, pH 7.5, 10âmM KCl, 1.5âmM MgCl2, 1âmM EDTA, 1âmM EGTA, 1âmM DTT, 1X cOmplete EDTA-free protease inhibitor cocktail (Roche) was added to the cells in being stirred in MIB buffer and poured into a nitrogen cavitation device kept on ice. The cells were subjected to a pressure of 500âpsi for 20âmin before releasing the nitrogen from the chamber and collecting the lysate. The lysate was clarified by centrifugation at 800âxâg and 4â°C, for 15âmin, to separate the cell debris and nuclei. The supernatant was passed through a cheesecloth into a beaker kept on ice. The pellet was resuspended in half the previous volume of MIBSM buffer (3 volumes MIB buffer + 1 volume SM4 buffer) and homogenized with a Teflon/glass Dounce homogenizer. After clarification as described before, the resulting lysate was pooled with the previous batch of the lysate and subjected to centrifugation at 1000âxâg, 4â°C for 15âmin to ensure complete removal of cell debris. The clarified and filtered supernatant was centrifuged at 10,000âxâg and 4â°C for 15âmin to pellet crude mitochondria. Crude mitochondria were resuspended in 10âmL MIBSM buffer and treated with 200 units of Rnase-free Dnase (Sigma-Aldrich) for 20âmin in the cold room to remove contaminating genomic DNA. Crude mitochondria were again recovered by centrifugation at 10,000âg, 4â°C for 15âmin and gently resuspended in 2âmL SEM buffer (250âmM sucrose, 20âmM HEPES-KOH, pH 7.5, 1âmM EDTA). Resuspended mitochondria were subjected to a sucrose density step-gradient (1.5âmL of 60% sucrose; 4âmL of 32% sucrose; 1.5âmL of 23% sucrose and 1.5âmL of 15% sucrose in 20âmM HEPES-KOH, pH 7.5, 1âmM EDTA) centrifugation in a Beckmann Coulter SW40 rotor at 28,000ârpm (139,000âxâg) for 60âmin. Mitochondria seen as a brown band at the interface of 32% and 60% sucrose layers were collected and snap-frozen using liquid nitrogen and transferred to â80â°C.
Frozen mitochondria were transferred on ice and allowed to thaw slowly. Lysis buffer (25âmM HEPES-KOH, pH 7.5, 50âmM KCl, 10âmM Mg(OAc)2, 2% polyethylene glycol octylphenyl ether, 2âmM DTT, 1âmg/mL EDTA-free protease inhibitors (Sigma-Aldrich) was added to mitochondria and the tube was inverted several times to ensure mixing. A small Teflon/glass Dounce homogenizer was used to homogenize mitochondria for efficient lysis. After incubation on ice for 5â10âmin, the lysate was clarified by centrifugation at 30,000âxâg for 20âmin, 4â°C. The clarified lysate was carefully collected. Centrifugation was repeated to ensure complete clarification. A volume of 1âmL of the mitochondrial lysate was applied on top of 0.4âmL of 1âM sucrose (v/v ratio of 2.5:1) in thick-walled TLS55 tubes. Centrifugation was carried out at 231,500âxâg for 45âmin in a TLA120.2 rotor at 4â°C. The pellets thus obtained were washed and sequentially resuspended in a total volume of 100âµl resuspension buffer (20âmM HEPES-KOH, pH 7.5, 50âmM KCl, 10âmM Mg(OAc)2, 1% Triton X-100, 2âmM DTT). The sample was clarified twice by centrifugation at 18,000âg for 10âmin at 4â°C. The sample was applied on to a linear 15â30% sucrose (20âmM HEPES-KOH, pH 7.5, 50âmM KCl, 10âmM Mg(OAc)2, 0.05% n-dodecyl-β-D-maltopyranoside, 2âmM DTT) gradient and centrifuged in a TLS55 rotor at 213,600âxâg for 120âmin at 4â°C. The gradient was fractionated into 50âμL volume aliquots. The absorption for each aliquot at 260ânm was measured and fractions corresponding to the monosome peak were collected. The pooled fractions were subjected to buffer exchange with the resuspension buffer to dilute away sucrose.
rRNA mass spectrometry
Purification of mitochondrial rRNAs
Mitochondrial rRNAs of the HEK293 cells were extracted from the purified ribosome using Isogen (Nippon Gene) and stored at â80â°C until use. TK6 cell culture (~1.0âÃâ109 cells) were homogenized by Dounce homogenizer with 250âmM sucrose, 1âmM EDTA and 10âmM TrisâHCl (pH 8.0). The lysate was centrifuged twice at 1500âxâg for 3âmin at 4â°C, and the resulting supernatant was further centrifuged at 5000âxâg for 10âmin to obtain mitochondrial precipitate. From the precipitate, total mitochondrial RNA (â~â100âμg) was extracted using TRIzol Reagent (Thermo Fisher Scientific).
Each mitochondrial rRNA was purified by using reversed-phase LC through a PLRP-S 4000âà column (4.6âÃâ150âmm, 10âμm, Agilent Technologies). After applying the mixture of rRNAs from the purified ribosome or total mitochondrial RNA to the column, the rRNAs were eluted with a 60-minute linear gradient of 11.5â13.5% (v/v) acetonitrile in 100âmM TEAA (pH 7.0) and 0.1âmM diammonium phosphate at a flow rate of 200âμl/min at 60â°C while monitoring the eluate at A26087.
LC-MS, MS/MS and MS/MS/MS analysis and database search of RNA fragments
RNA was digested as described below. Nucleolytic RNA fragments were analyzed with a direct nanoflow LC-MS system as described88. The LC eluate was sprayed online at â1.3âkV with the aid of a spray-assisting device88 to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) in negative ion mode. Other settings for MS, MS/MS and MS/MS/MS were as described in ref. 89.
Ariadne57 was used for database searches and assignment of MS/MS RNA spectra. The composite of human cytosolic and mitochondrial rRNA and tRNA sequences was used as a database. The following default search parameters for Ariadne were used: maximum number of missed cleavages, 1; variable modification parameters, two methylations per RNA fragment for any residue; RNA mass tolerance, ±5âppm, and MS/MS tolerance, ±20âppm. For assignment of Ψ residues using 5,6-D2-uridine labeled RNAs, the mass table and the variable modification parameters were altered from default values to â5,6D_CUâ and âΨâ, respectively, because both C and U were labeled with the medium and the pseudouridylation reaction results in the exchange of the position 5 deuterium of 5,6-D2-uridine to the proton of solvent, providing aâââ1âDa mass shift90.
Internal standard RNAs and SILNAS-based quantitation of the stoichiometry of post-transcriptional modification
To construct the plasmids for in vitro transcription of internal standard RNAs, DNAs encoding human 12âS and 16âS rRNAs were amplified by PCR from the mitochondrial DNA from TK6 cells. The PCR primers used are as follows: HindIII-12S-Forward TATAAAGCTTAATAGGTTTGGTCCTAGCCTTTCTATTAGC, 12S-BamHI-reverse ATATGGATCCGTTCGTCCAAGTGCACTTTCCAGTACACTT, HindIII-16S-forward TATAAAGCTTGCTAAACCTAGCCCCAAACCCACTCCACCT and 16S-XhoI-reverse ATATCTCGAGAAACCCTGTTCTTGGGTGGGTGTGGGTATA). The amplified DNAs were inserted into the multiple cloning sites of plasmid pCDNA3.1 (+) (Thermo Fisher Scientific). To synthesize RNA, 2âμg of template DNA was incubated and transcribed using Megascript T7 kit (Thermo Fisher Scientific). When RNA was synthesized, guanosine-13C10 5â²-triphosphate, cytidine-13C9 5â²-triphosphate, or uridine-13C9 5â²-triphosphate solution was used instead of the respective 5â²-triphosphate reagent that contained carbons with a natural isotope distribution. The RNA was precipitated in ethanol, solubilized in nuclease-free water, and purified further by reversed-phase LC as described above.
SILNAS-based quantitation was performed as described in ref. 91. In brief, RNA (â~â100 fmol) from natural sources or cells grown in guanosine with natural isotope distribution was mixed with an equal amount of synthetic RNA transcribed in vitro with 13C-labeled guanosine and digested with RNAse T1 (2âng/μl). For the RNA transcribed with 13C9-labeled cytidine and uridine, Rnase A (0.5âng/μl) was used as the digestion enzyme. Digestion was carried out in 100âmM TEAA (pH 7.0) at 37â°C. The 1:1 RNA mixing was performed based on the measurement of the absorbance at 260ânm and ensured later by a correction factor obtained experimentally. After obtaining the LC-MS spectrum of the digested RNA mixture, the stoichiometry of RNA modification at each site was estimated by Ariadne program designed for SILNAS91. The results were confirmed by manual inspection of the original MS spectrum to examine whether the estimates are based on âuncontaminatedâ MS signals.
Other procedures for RNA analysis
The masses of RNA fragments and a-, c-, w-, and y-series ions were calculated with Ariadne (http://ariadne.riken.jp/). Sequence-specific Rnase H cleavage of rRNAs was performed as described in ref. 92, using O-methylated RNA/DNA hybrid oligonucleotide complementary to the 12âS and 16âS rRNAs, GmUmUmCmGmUmCm(CAAG)UmGmCmAmCmUmUmUmCmCmAmGmUmAmCmAmCm and (AmAmCmCmCmUmGm(TTCT)UmGmGmGmUmGmGmGmUm, respectively (where Nm refers to 2â-O-methyl ribonucleotide and deoxyribonucleotides are indicated in parentheses).
SDS-PAGE and immunoblotting analyses
Whole-cell protein extracts were obtained by solubilization in RIPA buffer (25âmM Tris-HCl pH 7.6, 150âmM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS) with 1âmM PMSF and 1x mammalian protease inhibitor. Extracts were cleared by 5âmin of centrifugation at 10,000âg at 4â°C. Protein concentrations were determined with the Folin phenol reagent. 40â60âµg of whole-cell protein extracts were separated by SDS-PAGE in the Laemmli buffer system. Proteins were then transferred to nitrocellulose membranes, subsequently blocked with 5% skim milk, and then incubated with antibodies against the indicated proteins followed by a second reaction with anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase. Signals were detected by chemiluminescence and exposure to X-ray films. For quantification, the signals in digitalized images were quantified by densitometry using the histogram function of Adobe Photoshop. Values were normalized to the signal of β-Actin and plotted as the percentage of WT using the Prism 8 software.
Pulse labeling of mitochondrial translation products
Mitochondrial protein synthesis was determined by pulse-labeling of 60â70% confluent human wild-type (WT) HEK293T cells, MRPS29-KO HEK293T cells, and knockout cells stably reconstituted with mutant MRPS29 variants. Cultures were grown in a medium without methionine and in the presence of 100âµg/mL emetine to inhibit cytoplasmic translation as described in refs. 93,94. Cells were labeled for 15âmin at 37â°C with 100âµCi of 35S-methionine (PerkinElmer Life Sciences, Boston, MA). After incubation, the medium containing 35S-methionine was removed, the cells were washed once with PBS, and then the cells were incubated for 5âminutes in complete DMEM to allow recovery. After the short incubation, cells were washed once more with PBS, collected by trypsinization, and whole-cell protein extracts were obtained by solubilization with RIPA buffer. 40â50âµg of each sample was separated by SDS-PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and exposed to X-ray film. Membranes were then probed by immunoblotting to assess the steady-state levels of β-ACTIN as a loading control. Values were normalized to the signal of β-ACTIN and plotted as the percentage of WT using the Prism 8 software. Potassium cyanide (KCN)-sensitive endogenous cell respiration was measured polarographically using a Clark electrode in the indicated cell lines. Cell respiration rates are presented as nmol of O2 consumed per one million cells per minute.
Cryo-EM data acquisition
Five cryo-EM datasets of mitoribosomal complexes were collected in total using EPU 1.9 (Supplementary Table 1). First, 3âμL of ~120ânM mitoribosome was applied onto a glow-discharged (20âmA for 30âs) holey-carbon grid (Quantifoil R2/2, copper, mesh 300) coated with continuous carbon (of ~3ânm thickness) and incubated for 30âs in a controlled environment of 100% humidity and 4â°C. The grids were blotted for 3âsec, followed by plunge-freezing in liquid ethane, using a Vitrobot MKIV (FEI/Thermofischer). For high resolution mitoribosome analysis, five separate datasets were collected on FEI Titan Krios (FEI/Thermofischer) transmission electron microscope operated at 300âkeV, using C2 aperture of 70âμm and a slit width of 20âeV on a GIF quantum energy filter (Gatan). A K2 Summit detector (Gatan) was used at a pixel size of 0.83âà (magnification of 165,000X) with a dose of 29-32 electrons/à 2 fractionated over 20 frames. A defocus range of â0.6 to â2.8âμm was used. More detailed parameters for each one of the datasets are listed in Supplementary Table 1.
Cryo-EM data processing
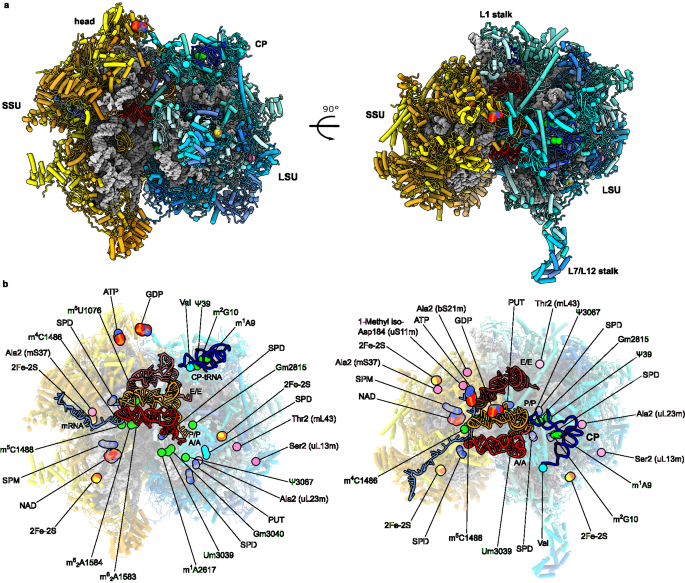
The complete workflows of the mitoribosomal complexes A/A-P/P-E/E, A/P-P/E, L1 stalk, monosome consensus map, SSU and their 8 masked regions are given in Supplementary Figs. 1 and 2. For all the datasets, beam-induced motion correction and per-frame B-factor weighting were performed for all data sets using RELION-3.0.236,37. Motion-corrected micrographs were used for contrast transfer function (CTF) estimation with gctf95. Unusable micrographs were removed by manual inspection of the micrographs and their respective calculated CTF parameters. Particles were picked in RELION-3.0.236,37, using reference-free followed by reference-aided particle picking procedures. Reference-free 2D classification was carried out to sort useful particles from falsely picked objects, which were then subjected to 3D classification. 3D classes corresponding to unaligned particles and LSU were discarded and monosome particles were pooled and used for 3D auto-refinement yielding a map with an overall resolution of 2.9â3.4âà for the five datasets. Resolution was estimated using a Fourier Shell Correlation cut-off of 0.143 between the two reconstructed half maps. Finally, the selected particles were subjected to per-particle defocus estimation, beam-tilt correction and per-particle astigmatism correction followed by Bayesian polishing. Bayesian polished particles were subjected to a second round per-particle defocus correction. A total of 994,919 particles from all datasets were then pooled and separated into 86 optics groups in RELION-3.138, based on acquisition areas and date of data collection. Beam-tilt, magnification anisotropy and higher-order (trefoil and fourth-order) aberrations were corrected in RELION-3.138. Subsequently, 509,691 particles from datasets 2-4 were pooled together and subjected to 3D auto-refinement giving a final nominal resolution of 2.21âà . Masked refinement was performed on the small subunit head, body, mS39 and tail to produce maps of 2.36âà , 2.31âà , 2.44âà and 2.45âà resolution, respectively and likewise for the SLU body, L10-L7/L12 stalk, CP and L1 stalk to yield maps of 2.08âà , 2.38âà , 2.36âà and 2.89âà resolution, respectively. To improve the quality of the L1-stalk map, 220,906 monosome particles with high occupancy of E/E-tRNA were pooled and subjected to partial signal subtraction using the mask that retains the E/E-tRNA and L1-stalk, followed by 3D auto-refinement to have a nominal resolution of 2.87âà (Supplementary Fig. 1 and Supplementary Table 1).
For separating the A/A-P/P-E/E and A/P-P/E states, all 994,919 monosome particles were subjected to signal subtraction using the mask that retains the tRNA binding region. The tRNA binding sites displayed weak densities corresponding to a heterogeneous mixture of tRNAs bound with partial occupancy. Signal subtracted particles were 3D classified without alignment using a mask on the A-site. This yielded two main classes, one with density in the A-site (332,187 particles) and the other lacking it (629,597 particles). Particles with density in the A-site were selected and subjected to a second round of 3D-classification using a mask around the P- and E-sites. Two classes were selected, one with P/P- and E/E-tRNA density and other in hybrid state with P/E-tRNA density. All 3D classifications with signal subtracted particles were performed using a T-value of 400. The subtracted signal was reverted for these two classes, corresponding to the A/A-P/P-E/E state (82,522 particles) and A/P-P/E state (20,143 particles). Following 3D auto-refinement, the nominal resolution of the cosmplexes was 2.63âà and 2.98âà , respectively. Masked refinement for the classical state was performed using local masks on SSU head, body, tail, and mS39 to produce maps at 2.72âà , 2.75âà , 2.91âà and 2.93âà resolution, respectively. For the LSU body, L10-L7/L12, CP, and the L1 stalk regions, the resolution was 2.44âà , 2.84âà , 2.78âà and 3.20âà , respectively. Masked refinement for the hybrid state was performed using the same local masks as for the classical state but transformed to compensate for the structural differences. This produced maps for the SSU head, body, tail, and mS39 to produce maps of 3.03âà , 3.07âà , 3.39âà and 3.47âà resolution, respectively. For the LSU body, L10-L7/L12 region, CP, and the L1 stalk masks, the resolution was 2.84âà , 3.29âà , 3.25âà and 3.53âà , respectively. The maps were then subjected to modulation transfer function correction, automatic B-factor sharpening and local resolution filtering using RELION-3.1 (Supplementary Fig. 2, Supplementary Table 1).
Model building and refinement
We started by building a model into the 2.21âà resolution consensus map. The starting model for the LSU was taken from PDB ID: 6ZSG, and for the SSU from PDB ID: 6RW4. These models were rigid body fitted using UCSF Chimera96. Individual local-masked refined maps with local-resolution filtering and B-factor sharpening (RELION 3.1) superposed to the overall map were combined into a single composite map using Phenix.combine_focused_maps97 for model building and refinement. Coot v0.998 with Ramachandran and torsion restraints was used for model building.
The L1-stalk model was built using the 2.87âà resolution masked-refined map of L1 stalk from E/E-tRNA-containing particles. Initial model for uL1m was generated by homology-modeling with uL1 structure of T. thermophilus (PDB ID: 3U4M) as template using the Swiss-Model webserver99. The N-terminal extension of uL1m (residues 61â93), the C-terminal domain (residues 158â254) and the preceding loop (residues 148â157) of uL9m, and H76-77 (residues 2761â2786) of 16âS rRNA were modeled manually into the density. Mitochondria-specific C-terminal extension of uL9m was modeled as a helix based on the density and secondary structure prediction by PSIPRED100. The model was rigid body fitted into the respective local-masked refined maps for L1 stalk region in the A/A P/P E/E and A/P/E states followed by self-restrained real space refinement in Coot v0.9.
In the CP, the tRNAVal could be completely modeled, owing to the improved resolution of the local masked-refined map (2.36âà ). The 3â²-CCA terminus (C74âA76) and a valine residue linked to A76 could be placed on the density map. The N-terminus of mL40 was extended by 13-residues (47â59) modeled in a tubular wedged between the A- and P-site tRNAs to the phosphate backbone of mRNA residue 7. Similarly, the C-terminus of bL31m was extended by 31 residues (95â125) from the C-terminal helix onto SSU head. The local resolution was sufficient for accurate side-chain modeling for most residues and secondary-structure assignments. We modeled a disulfide linkage between C45-C76 of mS37. However, they might exist in both disulfide-linked, and reduced states as indicated by the density and a water mediated hydrogen bond with a neighbouring Arginine side chain (R240 from uS7m). The other pair C55-C66 appears to be reduced. However, as we had added 1âmM DTT to our ribosome purification buffers, this is not truly indicative of the native state of C45-C76 or C55-C66 pairs.
At the mRNA channel entrance, a more accurate and complete model of mS39 could be built with 29 residues added to the structure. Improved local resolution enabled unambiguous assignment of residues to the density which allowed us to address errors in the previous model. Finally, a total of 28 α-helices could be modeled in their correct register and orientation. Further, a 28-residue long N-terminal loop of mS31 (residues 247â275) along mS39 and mitochondria-specific N-terminal extension of uS9m (residues 53â70) approaching mRNA were modeled manually by fitting the loops into the density maps.
Polyamines were identified upon a further manual inspection of the density map for the presence of continuous tubular densities which corresponded to spermine, spermidine or putrescine in length. A total of one spermine, four spermidines and one putrescine were placed in the model, justified by the electrostatic and hydrogen-bonding interactions that each polyamine exhibits at its amine groups with its neighbors.
Metal ions, Mg2+, K+ and Zn2+, were modeled into densities based on a manual inspection of small density blobs and the chemical environment, specifically, coordination geometry and strength of density in both unsharpened and local resolution filtered maps. The coordination distance of Mg2+ is around 2âà with an octahedral geometry, whereas that of K+ is around 3âà with more relaxed constraints on coordination geometry. Notably, a density that was modeled previously as Zn2+ coordinated by residues D224, D240, D241 and H93 of uS2m was assigned as a magnesium ion, indicated by the octahedral geometry of coordination with the uS2m residues mentioned above and two water molecules.
A-, P- and E-site tRNAs, and mRNA were modeled manually in the density. The density for tRNAs and mRNAs comes from a natively-bound heterogeneous population. Hence, individual residues were modeled as one of the nucleotides, A, U, G, or C, based on the density and/or conservation. While the 70 residues of P-site tRNA could be modeled, fewer residues could be modeled for the A-site (22 residues) and E-site tRNAs (43 residues), including the acceptor arm and anticodon loop due to weaker respective densities. Along the mRNA channel, 34 residues of mRNA were built. In the tRNA binding region, codon-anticodon pairs were clearly resolved. Additionally, extra residues near the A-site (10-12, 14-16) and at the mRNA channel entrance (25, 27-29) were modeled accurately into density. For the remaining mRNA chain at a relatively lower resolution, correct overall chain trace and nucleotide orientation could be assigned. This allowed specific protein-nucleotide interactions to be identified for uS5m, uS7m, uS9m, uS12m, mS35, mS39 and mL40.
For modeling the classical A/A P/P E/E- and hybrid A/P P/E-states, the LSU and SSU from the consensus model were fitted into their composite maps, followed by manual revision. As described above, tRNAs and mRNA were modeled manually into the density.
The water molecules were automatically picked by Coot v0.961, followed by manual revision. Geometrical restraints of modified residues and ligands used for the refinement were calculated by Grade Web Server (http://grade.globalphasing.org) or obtained from CCP4 library101. Hydrogens were added to the models except for water molecules by REFMAC5102 using the prepared geometrical restraint files. Charged α-amino-group hydrogens (H2 and H3) and a free-ribose hydrogen HO3Ⲡwere removed from non-terminal protein and RNA residues, respectively, while those at terminal residues were kept. Protonation of Histidine residues were judged based on the chemical environment and removed or kept their N-H hydrogens (HD1 and HE2) accordingly. Protonation/deprotonation of ligands and modified residues were also adjusted.
Final models were further subjected to refinement with Phenix.real_space_refine v1.13_299897, wherein three macro-cycles of global energy minimization with reference restraints (using the input model as the reference, sigma 5-7) and secondary structure restraints, rotamer restraints but without Ramachandran restraints were carried out for each model. The composite maps were used as the targets for the refinement. An âeditâ file that defines metal-coordination bonds was generated by ReadySet in the Phenix suite and used. Non-canonical covalent bonds, which are those between tRNAVal and Valine and between 1-methyl-isoaspartate 184 and G185 in uS11m, were also defined in the âeditâ file. Model refinement data are listed in Supplementary Table 1.
Bioinformatic analysis of the Y-N-C-Y motif
The density and base-specific interactions of mRNA residues 25-28 with mS39 imply a sequence of pyrimidine, any nucleotide, cytosine, pyrimidine, leading to a consensus motif: Y-N-C-Y. We explored the possibility of increased frequency of this motif in mRNA sequences. The sequences for all 13 mRNAs were obtained from GenBank. The consensus motif was searched in each sequence and all occurrences (including ones that overlap) were recorded and used to calculate the percentage frequency of occurrence for each transcript and the average percentage frequency. To determine if there is an enrichment of this motif in mRNAs, we calculated its occurrence in a set of nuclear-encoded mRNAs. To create this set, we acquired sequences randomly from GenBank and subjected them to a 90% redundancy cut-off, resulting in a total of 1387 sequences. The percentage frequency of occurrence of Y-N-C-Y motif was determined for each of the 1387 sequences and used to calculate an average percentage frequency for the entire set. Next to determine the distribution of the Y-N-C-Y motif across the length of mRNAs, each occurrence was plotted against its position number in the mRNA sequence. This distribution for all 13 mRNA sequences was plotted together for comparison.
Molecular dynamics simulations: modified force fields for simulating a truncated mL64 protein and modified interactions with protein uL1m
Baseline force field used for simulations of P/E formation
An all-atom multi-basin structure-based âSMOGâ model59,60 of the mitoribosome was used to simulate P/E formation. For this, we first constructed single-basin structure-based models for the pre- and post-translocation configurations (A/A-P/P and P/P-E/E). We then combined the two models, such that the composite model stabilizes both the pre- and post-translocation configurations. Here we describe the baseline model, which is described in the main text. In subsequent sections, we provide detailed descriptions of all modified force fields.
In a single-basin all-atom structure-based model, every non-hydrogen atom is represented by a bead of unit mass, and an experimentally-obtained structure is used to define the potential energy minimum. The functional form of the potential is given by equation (1):
$$U= {\sum}_{{{{{{\rm{bonds}}}}}}}\frac{{\epsilon }_{r}}{2}{({r}_{i}-{r}_{i,0})}^{2}+{\sum}_{{{{{{\rm{angles}}}}}}}\frac{{\epsilon }_{{{{{{\rm{\theta }}}}}}}}{2}{({{{{{{\rm{\theta }}}}}}}_{i}-{{{{{{\rm{\theta }}}}}}}_{i,o})}^{2}\\+ {\sum}_{{{{{{\rm{impropers}}}}}}}\frac{{\epsilon }_{\chi imp}}{2}{({\chi }_{i}-{\chi }_{i,0})}^{2}+{\sum}_{{{{{{\rm{planars}}}}}}}{\epsilon }_{\chi planar}[1-\,\cos (2{\chi }_{i})]\\+ {\sum}_{backbone\,dihedrals}{\epsilon }_{bb}F({{\phi }}_{i}-{{\phi }}_{i,0})+{\sum}_{{{{{{\rm{sidechain}}}}}}\,{{{{{\rm{dihedrals}}}}}}}{\epsilon }_{sc}F({{\phi }}_{i}-{{\phi }}_{i,0})\\+ {\sum}_{{{{{{\rm{contacts}}}}}}}{\epsilon }_{c}\left[{\left(\frac{{\sigma }_{ij}}{{r}_{ij}}\right)}^{12}-2{\left(\frac{{\sigma }_{ij}}{{r}_{ij}}\right)}^{6}\right]+{\sum}_{{{{{{\rm{non}}}}}}-{{{{{\rm{contacts}}}}}}}{\epsilon }_{nc}{\left(\frac{{\sigma }_{nc}}{{\sigma }_{ij}}\right)}^{12}$$
where
$$F({\phi })=[1-\,\cos ({\phi })]+\frac{1}{2}[1-\,\cos (3{\phi })]$$
r0 and θ0 parameters are assigned values found in the Amber ff03 force field103. The dihedral parameters Ï0 are defined by the experimental structure. Ï0 non-planar improper dihedrals (around chiral centers) are also defined to have the values adopted in the experimental structures. Planar dihedral angles are given a cosine term with periodicity 2 and minima at 0 and 180 degrees. The energy scale is set to \({\epsilon }_{r}=100\frac{\epsilon }{{A}^{2}},{\epsilon }_{\theta }=80\frac{\epsilon }{{{rad}}^{2}},{\epsilon }_{{\chi }_{{imp}}}=10\frac{\epsilon }{{{rad}}^{2}},{\epsilon }_{{\chi }_{{planar}}}=40\frac{\epsilon }{{{rad}}^{2}}\), where \(\epsilon\) is the reduced energy scale. The dihedral and contact energy scales \({\epsilon }_{{bb}},{\epsilon }_{{sc}},{\epsilon }_{C}\) are normalized as in ref. 59.This combination of structure-based non-bonded terms and AMBER bonded terms is identical to the implementation of a previous SMOG-amber variant (called SBM_AA-amber-bonds104 in the SMOG 2 force field repository (called SBM_AA-amber-bonds in the SMOG 2 force field repository at smog-server.org), though post-transcriptionally-modified residues were added for the current model. Upon publication, the SMOG 2 force field templates used in the current study will be available for download on the SMOG 2 force field repository (ID: AA_PTM_Hassan21.v1).
For the non-bonded interactions, each atomic contact that is found in the experimental structure (i.e. ânativeâ contact) is given an attractive Lennard-Jones-like interaction that stabilizes a preassigned structure. Native contacts were defined based on the Shadow Contact Map algorithm with default parameters105. Ïij are the interatomic distances between contacts in the native structure, multiplied by 0.96. As employed previously in simulations of the ribosome106,107 and viral capsids104, this scaling of the contact distance was employed to avoid artificial expansion of the ribosome that results from thermal energy. That is, while the potential energy is defined such that the experimental structure is the global minimum, thermal energy leads to a free-energy minimum in which the ribosome is slightly expanded. By shortening the stabilizing interactions, the free-energy minimum is consistent with the experimental structure. For this comparison, the radius of gyration (Rg) was used as a measure of expansion. With this scaling of the contact distances, Rg was found to match that of the cryo-EM model. All non-native contacts are given a repulsive term that ensures excluded-volume steric interactions are present. The parameter Ïnc is given the value 2.5âà , while \({\epsilon }_{{nc}}=0.1\epsilon .\)
Stabilizing intra-ribosome contacts and dihedrals were assigned the values found in the A/P-P/E (SSU rotated) hybrid structure, which is described in the main text. This ensures that the SSU rotates forward in each simulation. Each simulation is initiated from the A/A-P/P conformation. All intra-mt-tRNA and intra-mt-mRNA contacts were defined based on the A/A-P/P structure. Since stabilizing contacts in a structure-based model describe effective interactions108,109, we rescaled the strengths of subsets of contacts within the model. Consistent with recent simulations of P/E formation in bacteria106, interactions that stabilize the structure of the ribosome (i.e. intra-ribosomal) are given larger weights than contacts that are only transiently-formed (e.g. tRNA-ribosome contacts). Relative to intra-ribosome contacts, the energetic interactions between the mt-mRNA-tRNA complex and the SSU (in A/A-P/P and P/P-E/E structures) were rescaled by a factor of 0.3. While these interactions were weaker than intra-ribosomal contacts, since the simulations were not performed under conditions in which translocation will occur (e.g., binding of elongation factors or back-rotation, etc), the mt-mRNA maintained its initial position on the mRNA track of the SSU. P/P-E/E contacts between the mt-tRNA and the uL1m protein were rescaled by a factor of 0.8 and all other interactions between mt-mRNA-tRNA and the LSU were rescaled by a factor of 0.1. This rescaling was applied to account for the transient nature of mt-mRNA-tRNA binding to the ribosome. The strength of the uL1m-tRNA contacts was set such that the P/E configuration would stably form contacts with the uL1m protein. When contacts were scaled below a factor of ~0.5, we found that uL1m could not form stable interactions with the mt-tRNA.
Interactions between the 3â-CCA ends of A-site and P-site mt-tRNAs with the A-site and the P-site were scaled by a factor of 0.2, while their interactions with P-site and E-site were scaled by a factor of 0.8. This leads to the mt-tRNA molecules favoring binding of their target sites (i.e., sites bound in the P/E configuration) on the ribosome. While the interactions with the subsequent binding site (i.e., A-site mt-tRNA with the P-site and P-site mt-tRNA with the E-site) were stronger, these contacts are short range (~6âà ). Accordingly, the stronger weights do not affect the dynamics of the mt-tRNA until it is very close to the binding site. Thus, the details of the final repositioning of the 3â-CCA tail of the P-site mt-tRNA should not be overinterpreted. While the contacts are short-range, the structure of the mitoribosome may introduce steric barriers that the mt-tRNA must navigate in order to adopt the P/E configuration109. With these considerations in mind, the current model is aimed at identifying structural elements that can impose steric limitations on mt-tRNA during P/E formation.
There were also two sets of harmonic potentials that were introduced. To mimic the presence of a nascent peptide chain, which would restrain the 3â-CCA end of the A-site tRNA to the PTC, we introduced weak harmonic restraints (weight of 1/nm2) between the 3â-CCA end of the A-site tRNA and the P site of the LSU. Distances were assigned based on the P/P conformation. These weak interactions ensured that the 3â-CCA end remains localized to the PTC. We also introduced harmonic potentials between base-pairing contacts in the mt-tRNA acceptor arm (weight 1000/nm2). Due to the details of the contact map, initial simulations exhibited partial unfolding/refolding of the acceptor arm during P/E formation. Since that was likely an artifact due to the simplicity of the model and the details of the contact map definition, these additional harmonic terms were introduced to ensure the tRNA acceptor arm remained folded throughout the transition.
Modified force fields
Multiple variants of the model (i.e., force field) were considered, in order to investigate the influence of steric interactions between the P-site mt-tRNA and C-terminal residues of mL64 and to investigate the influence of attractive interactions between uL1m and P-site mt-tRNA on P/E formation.
Truncated-mL64 model: ÎmL64 force field
The C-terminal region of mL64 is involved in steric interactions with the mt-tRNA. For ÎmL64 model, the C-terminal region of mL64 (beginning with Q112) was removed from the system. Q112 was chosen as truncating here guarantees no steric interactions between mL64 and the mt-tRNA and keeps the rest of the protein intact. Thus, all other interactions were unperturbed. This model is intended to mimic an ideal experiment, where one could delete the terminal region of mL64 without introducing indirect effects on the dynamics. In our application, this model allows us to ask what effect steric interactions between P-site mt-tRNA elbow and mL64 have on the dynamics of P/E formation. Deletion of this region from the model was achieved using the smog_extract tool of the SMOG 2 software package59.
Modified-uL1m model: ÎuL1m force field
All stabilizing contacts between uL1m and the P-site mt-tRNA that are formed in the P/P-E/E state were scaled by a relative factor of 0.1, rather than 0.8 (baseline model, above). Thus, by weakening the attractive interactions, there is insufficient stabilizing energy between uL1m and the P/E tRNA, such that the tRNA elbow does not fully reach the P/E configuration.
Combined truncated mL64 and modified uL1m model: ÎmL64-ÎuL1m force field
All modifications applied in the truncated mL64 model and the modified-uL1m model were included in a single combined force field.
Simulation details
All simulations were initiated from the pre-translocation A/A-P/P configuration. All force field files were generated using the SMOG 2 software package59. Molecular dynamics simulations were performed using Gromacs (v5.1.4)110,111. Reduced units were used in all calculations. Energy minimization was first performed using steepest descent minimization. The system was then coupled to a heat bath using Langevin Dynamics protocols, with reference temperature of 0.5 É/kB reduced temperature units (60 Gromacs units). This temperature was chosen since it has been shown to produce structural fluctuations that are consistent with those inferred from anisotropic B-factors and explicit-solvent simulation112. The timestep was 0.002 reduced units and each simulation was continued until the distance of the 3â-CCA end from the LSU E-site RCCA reached a value less than 4âà . Energy minimization was initially performed using steepest descent minimization. Each simulation was performed for a maximum of 108 timesteps. For the primary/baseline model described in the main text (327 simulations) we obtained an aggregate simulated time of 2.27 *106 reduced units. For the truncated mL64 model (ÎmL64 model) (321 simulations), the aggregate simulation time was 1.87 *106 reduced units. For the attenuated uL1m contacts model (ÎuL1m model) (237 simulations), the aggregate simulation time was 1.76 *106 reduced units. For the ÎmL64-ÎuL1m model (181 simulations), the aggregate simulation time was 1.04 * 106. Previous comparisons of diffusion coefficients of tRNA molecules in the ribosome (explicit-solvent simulations vs. SMOG models), estimate that 1 reduced unit corresponds to an effective timescale of approximately 1 nanosecond113. Using this conversion factor, the presented simulations represent an aggregate effective simulate time of ~ 6.94 milliseconds.
Structural metrics for describing simulated dynamics
– A-site mt-tRNA, \({R}_{{CCA}}^{P}\): distance between the geometric centers of the side chains of C75 in the A-site mt-tRNA and Gm2815 in the LSU rRNA. These residues base pair in the A/P-P/E and P/P-E/E configurations.
– P-site mt-tRNA, \({R}_{{CCA}}^{P}\): distance between the geometric centers of the side chains of C75 in the P-site mt-tRNA and Gm2815 in the LSU rRNA. These residues base pair in the A/P-P/E configuration.
–RCCA: distance between the geometric centers of the side chains of A76 in the P-site mt-tRNA and G2909 in the LSU rRNA. G2909 in the E-site forms a base stacking interaction with A76 of P-site mt-tRNA in the A/P-P/E configuration.
–Relbow: distance between geometric centers of U54 of P-site mt-tRNA and its reference position in the A/P-P/E configuration after alignment of the LSU rRNA.
– \({Q}_{L1-{tRNA}}\): the fraction of P/P-E/E contacts between uL1m and P-site mt-tRNA. A contact is defined as formed if it is within a factor of 1.2 of the distance found in the P/P-E/E configuration.
SSU rotation and tilt angles
To follow the rotary motions of the SSU body with respect to the LSU, we calculated rotation and tilt angles for both the SSU body. For this, we first determined the corresponding E. coli numbering of the mitoribosome rRNA residues via STAMP alignment114 of the mitoribosome to a reference E. coli structure (PDB ID: 4V9D, [https://doi.org/10.2210/pdb4V51/pdb]). Alignment was performed separately for the LSU, SSU head and SSU body. The angles were then calculated, using the assigned E. coli numbering.
Structural model preparation for simulations
Since the cryo-EM models did not resolve the exact same set of atoms, there was a need to perform some additional structural modeling steps prior to performing simulations. For example, to define the multi-basin structure-based model, it was necessary for each mt-tRNA molecule to have identical atoms in the pre- and post-translocation configurations. Additionally, to define the intra-ribosome interactions to stabilize the A/P-P/E conformation, it was necessary that all mt-rRNA and mitoribosomal proteins had identical atomic composition. While most of the resolved atoms are common to all reported models, we employed restraint-based modeling to construct models of the mitoribosome and mt-tRNA that have identical compositions. Below, we describe the steps required for modeling the mitoribosome and mt-tRNA.
Structural modeling pre-processing steps for rRNA and proteins for simulations
Since the structure-based model only included intra-ribosome interactions from the A/P-P/E structure, we first removed all mt-tRNA and mt-mRNA from the A/A-P/P and A/P-P/E models. We also removed ligands and water molecules (HOH, SPD, PUT, NAD, SPM). For the remaining atoms, we identified all atoms that were common to the A/A-P/P and A/P-P/E structures. We then generated a single-basin structure-based model of the A/A-P/P structure. Next, position restraints (harmonic terms, weight of 100 for each atom) were introduced for all common atoms, with positions corresponding to the A/P-P/E structure. The system was then energy minimized via steepest descent minimization. Then, the system was subject to simulated annealing. The system was initially simulated at 0.15 reduced temperature units for 25,000 timesteps. The temperature was then linearly decreased to 0 over the next 50,000 timesteps. An additional 25,000 steps were simulated at temperature 0. The final, annealed structural model was used for force field generation steps, as described above.
Structural modeling pre-processing steps for mt-tRNA and mt-mRNA
Since the cryo-EM reconstructions reported consensus/average sequences for each mt-tRNA, the composition of the A-, P- and E-site mt-tRNAs were not identical. To perform simulations, we homogenized the composition of the mt-tRNA molecules in the A/A-P/P-E/E structure and the mt-tRNA codons. For this, we started with the structural model of the A-site mt-tRNA (and the associated codon) from the A/A-P/P structure, and then used it to generate a structure-based model. We then introduced position restraints based on the positions of the A-site mt-tRNA in the A/A-P/P-E/E structure. All common atoms (i.e., backbone atoms, or atoms in identical residues) were restrained to the A-site mt-tRNA/mRNA positions. For the A-site mt-tRNA, restraints were not imposed on residues 46â58 and 16â17. Cycles of minimization and annealing were performed, where the position restraints were given weights of 1, 10, 100 and then 1000. This process was repeated for the P-site and E-site mt-tRNAs, as well as for the upstream and downstream mt-mRNA. This process produced a model of the A/A-P/P-E/E configuration that has repeating codons and identical mt-tRNA composition. This structural model was used to define the pre-translocation and post-translocation structure-based models, which were used to construct the multi-basin model.
Describing the sequence of rearrangements in simulations
To describe the dynamics of the P-site mt-tRNA during P/E state formation, we have defined three states based on the 2D probability distribution of \({R}_{{elbow}}^{E}\) and \({R}_{{CCA}}^{E}\) distances (Fig. 8). These states are defined as all configurations of the P-site mt-tRNA in the baseline model that sample the following values of \({R}_{{elbow}}^{E}\) & \({R}_{{CCA}}^{E}\):
state I1: disk centered at (\({R}_{{elbow}}^{E}\),\({R}_{{CCA}}^{E}\)) = (46,35) à with radius râ=â6âÃ
state I2: disk centered at (\({R}_{{elbow}}^{E}\),\({R}_{{CCA}}^{E}\))â=â(63,23) à with radius râ=â8âÃ
state I3: disk centered at (\({R}_{{elbow}}^{E}\),\({R}_{{CCA}}^{E}\)) = (42,13) à with radius râ=â7âÃ
Using this definition of the intermediate states, the most common sequence of transitions followed the order P/Pâ=â>âI1â=â>âI2â=â>âI3â=â>âP/E.
Structure analysis and figures
Visualization and analysis of the models and maps was carried out using UCSF Chimera and ChimeraX96,115. For model and map comparisons, models were superposed in Coot v0.961 using the Secondary Structure Matching algorithm and maps downloaded from EMDB were resampled on our maps in UCSF Chimera. Simulated snapshots were visualized using Visual Molecular Dynamics116.
Statistics and reproducibility
All experiments were done in at least biological triplicate unless otherwise stated. GraphPad Prism 8 software was used for statistical analyses of the data. One-way analysis of variance (ANOVA) was performed, comparing the mean of each cell line with the mean of the mS29-KO cell line reconstituted with WT mS29, followed by Dunnettâs multiple comparisons test. The data is presented as the meanâ±âSD; * Pââ¤â0.05, ** Pââ¤â0.01, *** Pââ¤â0.001, **** Pââ¤â0.0001. For immunoblotting, band density is quantified and normalized to ACTIN. For metabolic labeling of mitochondrially translated peptides in whole cells, the data presented represents the mean of each sampleâs 35S signals.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.