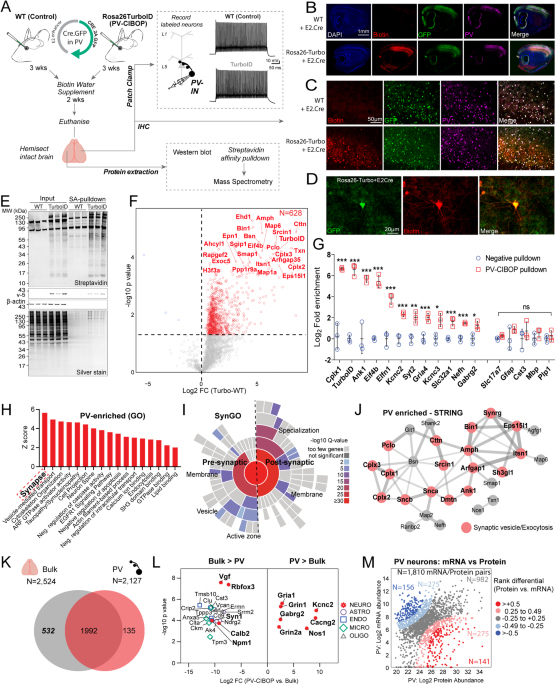
Native-state proteomics of PV interneurons in wild-type mouse brain
Proteomic biotinylation of native-state PV-INs (PV-CIBOP) was achieved by retro-orbital (RO) delivery of PV-IN-specific enhancer-targeting AAV (PHP.eB-E2-Cre-2A-GFP)33,40,41 into Rosa26TurboID/wt (PV-CIBOP group) or wild-type (WT) mice (Fig. 1A)16. For acute slice electrophysiology of PV-INs, we co-injected (RO) an AAV construct containing a floxed TdTomato sequence to fluorescently-label PV-INs due to non-visualization of GFP in ex vivo slices. After 3 weeks of Cre-recombination and 2 weeks of biotin supplementation8, we performed acute slice current clamp recordings confirming selective targeting and unaltered physiology of fast-spiking PV-INs by PV-CIBOP (Fig. 1A, Supplementary Fig. 1). To assess potential impacts of PV-specific TurboID expression and proteomic biotinylation on PV neuron function and overall local circuit activity, we obtained voltage and current clamp recordings from layer 5 pyramidal neurons and PV interneurons, respectively, in both WT control and PV-CIBOP mice (Supplementary Fig. 1A, B, H). Fast, spontaneous excitatory and inhibitory synaptic events (EPSCs and sIPSCs) were isolated during pyramidal cell recordings (Supplementary Fig. 1B). The amplitude, frequency and kinetic properties of both sEPSCs and sIPSCs were unperturbed in PV-CIBOP brains (Supplementary Fig. 1CâG). These results indicate that PV-CIBOP does not affect basal circuit excitability, synaptic receptor distributions or synaptic properties. In the same slices, neighboring TdTomato+ neurons exhibited fast, non-accommodating firing with narrow action potentials (Supplementary Fig. 1HâJ) and passive properties characteristic of fast-spiking PV-INs (Supplementary Fig. 1KâM). No differences in AP firing, various biophysical features, or passive properties were observed in PV-INs comparing PV-CIBOP with WT controls (Supplementary Fig. 1IâM). Immunohistochemical (IHC) studies from fixed PV-CIBOP cortices showed widespread biotinylation of PV-positive (PVâ+â) and GFP-positive neurons (Fig. 1B, C), in somatic and axo/dendritic compartments of PV-INs (Fig. 1D) without off-target biotinylation or reactive gliosis (Supplementary Fig. 2). Western blots (WB) showed strong biotinylation of a wide array of proteins in PV-CIBOP mice compared to few endogenously biotinylated proteins in WT control lysates (Fig. 1E)8. Using the N-terminal V5 (V5-TurboID) as a surrogate of Cre-mediated TurboID expression, we detected V5 in PV-CIBOP mice but not WT controls (Fig. 1E).
A Experimental outline to achieve native-state proteomics of PV-INs by CIBOP. BâD Immunohistochemistry of fixed brain sections confirmed biotinylation (red) of PV-INs (Pvalb: green) in PV-CIBOP but not in control mice (B: 4x and C: 20x magnification; D: Higher magnification (60x) images show biotinylation in PV-INs). E Top: Western Blot (WB) of input and streptavidin affinity pulldown samples confirms strong protein biotinylation in PV-CIBOP as compared to limited biotinylation in control animals. Bottom: Silver-stained gels of inputs and pulldown samples corresponding to WB images above. F Volcano plot representation of differential abundance of biotinylated protein, from PV-CIBOP and control mice. Red dots represent proteins biotinylated in PV-INs as compared to control mice (unpaired two-tailed T-test pââ¤â0.05). G Top PV-enriched proteins are shown on the left (including TurboID, Cnk1, Kcnc2, Kcnc3, Erbb4, Slc32a1 and GABA-ergic proteins). In contrast, non-neuronal (Mbp, Gfap, Aldh1l1, Cotl1) and excitatory neuronal (Slc17a7) proteins were not enriched (Data are presented as mean + SD, nâ=â3/group; unpaired two-tailed T-test *pâ<â0.05, **pâ<â0.01, ***pâ<â0.005). H Gene Ontology (GO) analyses of PV-enriched proteins compared to whole brain show enrichment of synaptic vesicle, GTPase binding, cytoskeletal and cell-projection related proteins. I SynGO analysis of PV-enriched proteins reveals labeling of proteins in pre- and post-synaptic compartments. J STRING analysis of PV-enriched synaptic proteins (>4-fold enriched over control) involved in synaptic vesicle and exocytosis, including complexins, ankyrins, synucleins. K Venn Diagram representing degree of overlap between proteins enriched in PV neurons, with whole brain proteomes from matched animals. L Top proteins differentially enriched in PV-INs as compared to the whole brain bulk proteome and those enriched in the bulk as compared to PV-INs, are highlighted. M Analysis of protein vs mRNA concordance in PV-INs, using PV-enriched proteins identified by PV-CIBOP and existing single nuclear transcriptomic data from mouse PV-INs. Based on differentials in rank abundances (protein vs. mRNA), discordant and concordant protein/mRNA pairs are highlighted. Also see Supplementary Figs. S1, S2, S3 and Supplementary Data 1 for related analyses and datasets. Source data are provided as a Source Data file. Image was created using BioRender.com.
Biotinylated proteins from PV-CIBOP and WT control samples were enriched using streptavidin (SA) beads followed by silver stain and WB (Fig. 1E), confirming enrichment of biotinylated proteins from PV-CIBOP mice, mirroring patterns observed in bulk brain lysates (inputs). Label-free quantitative MS (LFQ-MS) of SA-enriched samples identified a PV-IN proteome of 628 proteins enriched in PV-CIBOP samples as compared to controls (ââ¥â2-fold enriched and unadjusted pââ¤â0.05; 192 proteins â¥2-fold enriched at the FDRââ¤â0.05 threshold) (Fig. 1F, Supplementary Data 1). These included canonical PV-IN proteins, including Kv3 channels (Kcnc1, Kcnc2, Kcnc3; which were expressed at similar levels relative to each other), Gria4, Syt2 and Ank1, while markers of excitatory neurons (Slc17a7), astrocytes (GFAP), microglia (Cst3), oligodendrocytes (Mbp, Plp1) were not enriched (Fig. 1G)33,42,43,44. Gene set enrichment (GSEA) (Fig. 1H) and SynGO (Fig. 1I)45 analyses of the PV-IN proteome showed over-representation of gene ontologies (GO) including synaptodendritic and axonal localization, neurotransmission, vesicle function, synapse organization, ARF-GTPase signaling, growth factor receptor signaling, tauopathy/synucleinopathy, and included pre- and post-synaptic compartments. Some of the most abundant synaptic PV-IN proteins were involved in synaptic vesicle trafficking, fusion and exocytosis and included complexins (Cplx 1-3), synucleins alpha and beta (Snca, Sncb), Bin1 and amphiphysin (Fig. 1J)46. In contrast with bulk brain proteomes (Fig. 1K, L), PV-IN-specific proteomes including GABAergic and Kv3 channels, proteins involved in synaptic vesicle function, endocytic/endosomal pathways, GTPase binding proteins, cytoskeletal proteins (Ank1)43,47 and ribosomal large subunit proteins (Rpl15, Rpl18, Rpl24). Non-PV-IN proteins and glial proteins were preferentially enriched in the bulk brain proteome (Fig. 1L). Using a reference dataset of brain protein half-lives generated by metabolic labeling48, we found that the PV-CIBOP proteome had a similar distribution of short and long half-life proteins (Supplementary Fig. 3). We also verified that the distribution of neuronal soma and axon/synaptic/dendrite-localized proteins in the PV-CIBOP proteome and bulk brain proteome, were similar (Supplementary Fig. 4). These analyses suggest that the PV-CIBOP proteome is indeed representative of the neuronal proteome, rather than being biased by protein turn-over or biased towards specific neuronal compartments. Lastly, we contrasted our PV-IN proteome with reference single cell/nuclear RNAseq PV-IN transcriptomes from mouse brain (Fig. 1M, Supplementary Fig. 5)49, revealing modest concordance between 1,810 mRNA-protein pairs (Spearmanâs Rho=0.27, pâ<â0.001) (Supplementary Data 1).
In summary, our PV-CIBOP studies successfully identified native-state proteomic signatures of PV-INs many of which are not captured by bulk brain proteomics and are discordant with mRNA-level findings.
Proteomic signatures of PV-INs in contrast to Camk2a excitatory neurons reveal molecular signatures associated with vulnerability and cognitive resilience
We contrasted proteomic signatures of PV-INs with Camk2a-positive excitatory neurons using the CIBOP approach in two independent cellular contexts (Fig. 2A)8. Of 1,841 proteins enriched in either PV-CIBOP or Camk2a-CIBOP proteomes (Fig. 2A, Supplementary Data 2), 1,578 were enriched in Camk2a neurons and 1,408 proteins enriched in PV-INs, with 1,135 proteins enriched in both. 245 proteins were highly-enriched (â>â4-fold) in PV-INs (including Kv3 channel proteins) while 163 proteins were highly-enriched in Camk2a neurons (Fig. 2B). Ribosomal, GABA metabolism, ephrin B pathway, clathrin-coated vesicle, transport, cytoskeleton, endoplasmic reticulum, calcium binding, synaptic vesicle exocytosis and Akt/mTOR signaling were over-represented in PV-IN-enriched proteins (Fig. 2C, Supplementary Fig. 6). In contrast, cellular metabolism, fatty acid oxidation, NAD binding, lipid metabolism, proteasome complex, ER-phagosome and mitochondrial terms were over-represented in the Camk2a-CIBOP-enriched proteome (Fig. 2C, Supplementary Fig. 6). Upstream analyses identified potential microRNA (miRNA) regulators of PV-INs and Camk2a neurons, including enrichment of microRNAs 133a and 133b targets in the PV-IN proteome (Supplementary Fig. 6), in agreement with prior miRNA tagging and affinity purification (miRAP)50 studies that identified miRNAs 133a and 133b as highly expressed in PV-INs. Two microRNAs recently identified as predictors of cognitive decline in humans (miR-29a and miR-132), were also predicted to regulate PV-IN proteomic signatures51. These analyses suggest that molecular signatures that define PV-INs may be regulated by distinct sets of miRNAs, some of which have known associations with cognitive decline in humans.
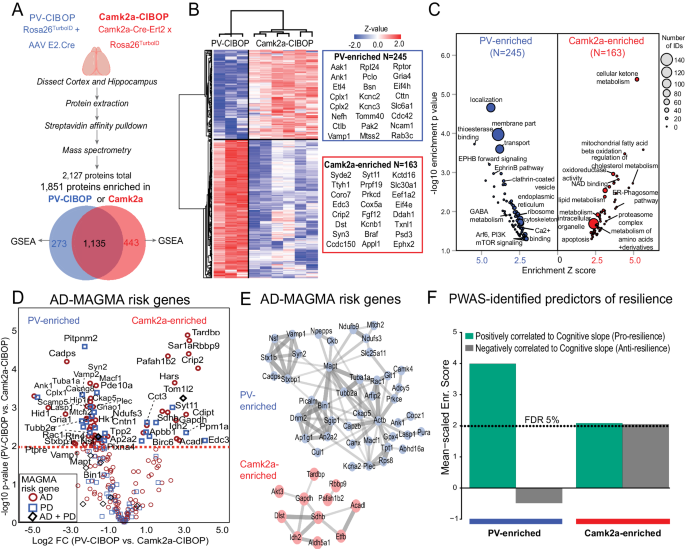
A Experimental outline for comparative analysis of CIBOP-based proteomics of PV-INs and Camk2a excitatory neurons from the mouse cortex by label-free quantitation MS analyses. 1851 proteins were quantified above negative samples in either PV-INs or Camk2a neurons. B DEA comparing PV-CIBOP (nâ=â3) and Camk2a-CIBOP (nâ=â6) cortex proteomes identified proteins >3-fold differential enrichment (signature proteins of each neuronal class), which were then hierarchically clustered. Top proteins (based on fold-change) are shown alongside the heatmap. C GSEA of PV-IN (blue) and Camk2a (red) signature proteins identified over-represented terms (GO, KEGG, Wikipathways, Reactome, Pathway commons) for PV-IN and Camk2a neurons. X-axis represents enrichment Z score for a given term, and Y-axis represents level of statistical significance of enrichment (Fisher exact test). Size of each data point indicates the number of protein IDs in that enrichment term. D Volcano plot representation of PV-IN and Camk2a neuron signature proteins which have known genetic risk associations in Alzheimerâs disease (AD) and Parkinsonâs disease (PD) based on MAGMA. Some proteins have shared genetic risk associations with AD and PD (Y-axis: Camk2a-CIBOP vs. PV-IN-CIBOP unpaired two-tailed T-test *pâ<â0.05). E Protein-protein interaction network (STRING) of AD-associated MAGMA risk genes with differential enrichment in PV-INs and Camk2a neurons. Clusters of mitochondrial, synaptic vesicle and endocytosis related proteins were revealed in PV-IN AD MAGMA risk genes. F Enrichment of PWAS-identified proteins associated with cognitive slope in PV-enriched and Camk2a-enriched proteomic signatures. Cognitive slope was estimated in ROSMAP cases. Positive slope indicates cognitive stability or resilience while a negative slope indicates cognitive decline. Proteins positively correlated with cognitive slope are referred to as pro-resilience proteins while those negative correlated with cognitive slope are anti-resilience proteins. Enrichment of pro-resilience and anti-resilience proteins in PV-enriched and Camk2a-enriched proteins identified by CIBOP was assessed after weighting based on strength of association between proteins and cognitive slope. FDR 5% threshold is shown. Also see Supplementary Fig. 4 and Supplementary Data 2 for related analyses and datasets. Image was created using BioRender.com.
To identify neurodegeneration-relevant proteins in PV-INs, we cross-referenced PV-IN and Camk2a CIBOP proteomic markers with neurodegeneration-associated risk genes from Multi-marker Analysis of GenoMic Annotation (MAGMA) analyses (Fig. 2D, Supplementary Data 2)52,53. We identified 60 PV-IN AD-risk proteins related to synaptic vesicle fusion, docking and recycling (Bin1, Picalm, Dnm2, Ap1g1, Ap2a2, Sgip1), cytoskeleton and microtubules (Ank1, Actb, Tubb2a, Mapt), mitochondria (Mtch2, Ndufs3, Ndufb9, Slc25a11), and SNARE complex (Syn2, Stx1b, Vamp1, Nsf, Stxb1) (Fig. 2E). In comparison, 24 Camk2a neuron-enriched AD-risk proteins were identified, including oxidoreductases (Sdhb, Idh2, Aldh5a1, Etfb and Acadl), serine/threonine kinase Akt3 and TAR DNA binding protein (Tardbp).
We also leveraged data from recent protein-wide association studies of post-mortem human brains from participants in the Religious Orders Study and the Rush Memory and Aging Project (ROSMAP) longitudinal study in which post-mortem brain proteins correlated with rate of cognitive decline, were identified. Using cognitive slope as the outcome, proteins positively correlated with cognitive stability (a positive cognitive slope) represented pro-resilience proteins (nâ=â645). Conversely, proteins that were negatively correlated with cognitive stability (a negative cognitive slope), represented anti-resilience proteins (Nâ=â575)54,55. As compared to the Camk2a-CIBOP proteome, the PV-IN proteome was significantly enriched in pro-resilience proteins, including complexins (Cplx1, Cplx2), Ank1, highly-abundant PV-IN proteins (e.g. Aak1, Cttn, Bin1, Elfn1, Bsn) as well as ribosomal, mitochondrial, GTP binding, synaptic compartment and vesicle fusion proteins (Fig. 2F). While PV-CIBOP is selective to the PV-INs, Camk2a-CIBOP broadly labels several classes of excitatory neurons in the cortex56,57, which limits the ability to detect molecular characteristics of distinct excitatory neuronal sub-classes. Therefore, future studies should apply more selective AAV or transgenic approaches to contrast distinct excitatory neuronal sub-classes.
This evaluation of PV-IN proteomes, in contrast to Camk2a neurons, reveal a generalized molecular phenotype of high translational, synaptic vesicle transport and fusion (neurotransmission), GTP binding and signaling (Akt/mTOR) activities in PV-INs, including many AD-related genetic risk factors and proteins associated with cognitive resilience.
Network analyses of human post-mortem brain proteomes identify unique associations of PV-IN markers with neuropathology and cognitive dysfunction in AD
Supporting evidence for unique vulnerability of PV-INs to neurodegenerative disease pathology in AD may be revealed by analyses of bulk brain proteomes. We interrogated published human post-mortem bulk brain proteomic studies58,59,60,61,62,63,64,65,66,67 which included >500 post-mortem dorsolateral pre-frontal cortex samples (non-disease controls, Asymptomatic AD, and AD with dementia cases) from the ROSMAP and Banner Sun Health cohorts66,67. Over 8,000 proteins quantified in this previous study coalesced into 44 groups of highly co-expressed proteins (modules, annotated as M1-M44)68, some of which were enriched in markers of distinct brain cell types (eg. M1, M5, M10, M33 for neurons, M11 and M12 for astrocytes, M11 and M21 for microglia, M3 for oligodendrocytes), as well as distinct molecular mechanisms (eg. MAPK signaling in module M7)68. These modules also have unique correlations to amyloid burden (CERAD), neurofibrillary tangles (Braak stage), and cognitive function (MMSE or global cognitive function) (Fig. 3A)58,67. Therefore, these modules can be treated as groups of proteins representative of distinct disease mechanisms, some of which may be cell type-specific.
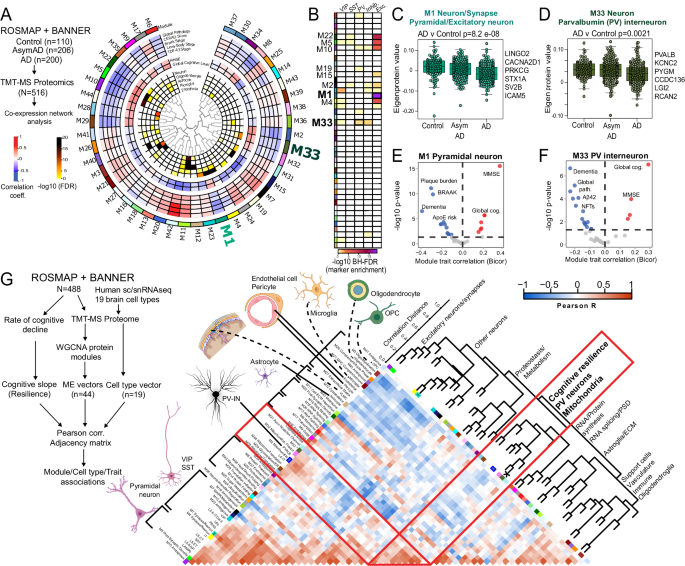
A Summary of network-based analysis of human bulk brain proteomes derived from post-mortem frontal cortex samples from controls, AsymAD and AD cases, from ROSMAP and BANNER cohorts (adapted from Johnson et. al.)70. Protein co-expression modules (M1-42) are arranged in a circular manner. Dendrogram indicates module inter-relatedness. Module trait associations are arranged in layers: Cell type signatures with cell type-enrichment statistical significance (-log10 FDR; module-cognitive trait associations (MMSE and Global cognitive score); module-neuropathological trait associations (red: positive, blue: negative). B Modules that showed over-representation distinct neuronal markers (pan-excitatory, pan-inhibitory and IN classes, namely PV-IN, SST-IN and VIP-IN) are shown. Color indicates level of statistical significance of enrichment. C, D Comparisons of module eigenprotein abundances of M1 (C, pan-excitatory neuronal module) and M33 (D, PV-IN module) across controls, AsymAD and AD cases. Overall ANOVA p-value is shown and top neuronal class-specific proteins representative of M1 and M33 are highlighted. E, F Volcano plot representations of Module-trait correlations (X-axis: Bicor, Y-axis: -log10 p-value of correlation) for M1 (E) and M33 (F). Top correlated traits (including ApoE genetic risk based on allelic combinations of ApoE ε2, 3 and 4) are labeled. G Adjacency matrix analysis based on correlations between protein co-expression modules (ME vectors), Cell type abundance vectors (based on selected markers of 19 distinct brain cell types identified by sc/snRNAseq) and cognitive slope till time of death (Left: overall outline of the analytical plan). Right: Heatmap representation of Pearsonâs correlation-based adjacency matrix. Cell type vectors and module ME vectors with representative ontologies of each ME, are shown on the left of the heatmap. Dendrogram on the right indicates relatedness among ME vectors, cell type vectors and cognitive slope, and revealed a cluster of PV-IN module M33, PV-IN vector, mitochondrial module M2 and M32, and cognitive slope (see Supplementary Data 3 for additional details). See Supplementary Data 3 and Supplementary Fig. 7 for related analyses. Image was created using BioRender.com.
We analyzed this human post-mortem brain proteomic dataset, now using sets of neuron sub-class-specific markers, to identify protein modules that may represent disease mechanisms associated with distinct neuronal classes. We derived 1,040 genes with â¥4-fold enrichment in specific neuronal classes (glutamatergic pyramidal neurons and GABAergic neurons including PV, Sst, VIP subtypes) from the Allen brain single cell/nuclear RNAseq (sc/snRNAseq) atlas (Supplementary Data 3). Pan-excitatory/glutamatergic markers (Camk2a, Slc17a7) were enriched in modules M1, M5, M22, M10 and M4. Pan-inhibitory markers (Gad1-2, Slc32a1) were enriched in modules M33 and M23. Among the inhibitory neuron modules, M33 showed enrichment in PV-IN markers, including Pvalb and Kcnc2 [Kv3.2]), while VIP interneuron markers were enriched in M23 (Fig. 3B). Therefore, M1 (as well as M22, M5, M10 and M4) is enriched in cellular mechanisms of AD in excitatory neurons, while M33 represents AD patho-mechanisms impacting PV-INs. Both pan-excitatory M1 and PV-IN M33 module abundances were lower in AD cases (Fig. 3C, D) and were associated with cognitive function (MMSE and global cognitive function) at last clinical visit prior to death, and negatively associated with severity of dementia and neuropathology (both Aβ and tau) (Fig. 3E, F) although APOE genetic risk was associated with M1, but not M33. This analysis shows that markers of distinct neuronal sub-classes are indeed enriched in specific protein modules, some of which have correlations with clinical and pathological traits and are therefore likely to be pathologically relevant.
We next assessed whether excitatory and inhibitory neuronal signatures obtained from bulk brain proteomes, are associated with rate of cognitive decline (cognitive slope) in longitudinal studies of aging and AD55,69 (Fig. 3G and Supplementary Fig. 7A, B). Using consensus marker lists of cell types from reference human brain single cell/nuclear RNAseq studies, we applied single-sample GSEA (ssGSEA) to proteomic data from 488 post-mortem human dorsolateral pre-frontal cortex (DLPFC) samples from ROSMAP/BANNER studies, to estimate abundances of 19 classes of excitatory and inhibitory neurons, glia and vascular cells (Fig. 3G)1,70,71,72. We also assessed whether protein co-expression modules, some of which are specific to neuronal sub-classes, also correlated with cognitive slope. The associations between different cell type abundances, protein modules and rate of cognitive decline were assessed using a correlation-based adjacency matrix. We found that the PV-IN cell type abundance, modules M33 (enriched in PV-IN markers), M2 and M32 (mitochondria) as well as cognitive slope (cognitive resiliency) were highly inter-correlated (Fig. 3G). In addition to PV-INs, the layer 4 IT neuronal estimate was also positively correlated with cognitive slope (Supplementary Fig. 7A, B). The abundances of these two cell types are therefore linked to cognitive resilience. The association between PV-IN module M33 and cognitive resilience remained significant after adjusting for several co-existent neuropathologies (including amyloid, tau, synuclein), suggesting that the relationship between PV-IN abundance and cognitive resilience may be independent of neuropathology (Supplementary Fig. 7C)55.
Overall, these network-based and correlative analyses of bulk brain proteomic data indirectly suggest that distinct neuronal sub-classes may be differentially impacted in AD pathology, and support the idea that integrity of PV-INs and/or their proteins, may be determinants of cognitive resilience in AD.
Neuron-specific molecular signatures resolve vulnerability of PV neurons to progressive Aβ pathology in the 5xFAD mouse model
The cross-sectional nature and post-mortem sampling of tissue in human post-mortem brain proteomic studies, limit the ability to resolve the impact of aging and disease progression on neuronal protein changes in AD. Since these can be better assessed in disease models, we examined bulk brain proteomes from a mouse model of Aβ pathology. We analyzed TMT-MS data from 43 WT and 43 5xFAD mice (age span 1.8-14.4 mo., 50% male, 50% female), in which over 8,500 proteins were quantified (Fig. 4A, Supplementary Data 4). As expected in 5xFAD mice, age-dependent increase in Aβ pathology was associated with concomitant increase in levels of Apoe, microglial proteins (Trem2, Msn, C1qb) and astrocyte proteins (Gfap) (Fig. 4A). Using 10 and 14 month timepoints, we assessed whether proteomic changes at more established and advanced stages of Aβ pathology in 5xFAD mice, are representative of changes occurring in post-mortem human AD brain using proteomes (control, AD and AsymAD) from the published resource also used in Fig. 368. We observed proteome-wide moderate positive correlations in AD pathological changes occurring across species (Supplementary Fig. 8AâC). Mechanisms involving splicing, proteasome, innate immunity, actin-cytoskeleton and MAPK signaling were increased in human AD and mouse 5xFAD brains (Supplementary Fig. 8D). Conversely, mitochondrial respiration, mitochondrial translation, endocytosis and synaptic signaling were decreased in human AD and mouse 5xFAD brains (Supplementary Fig. 8E).
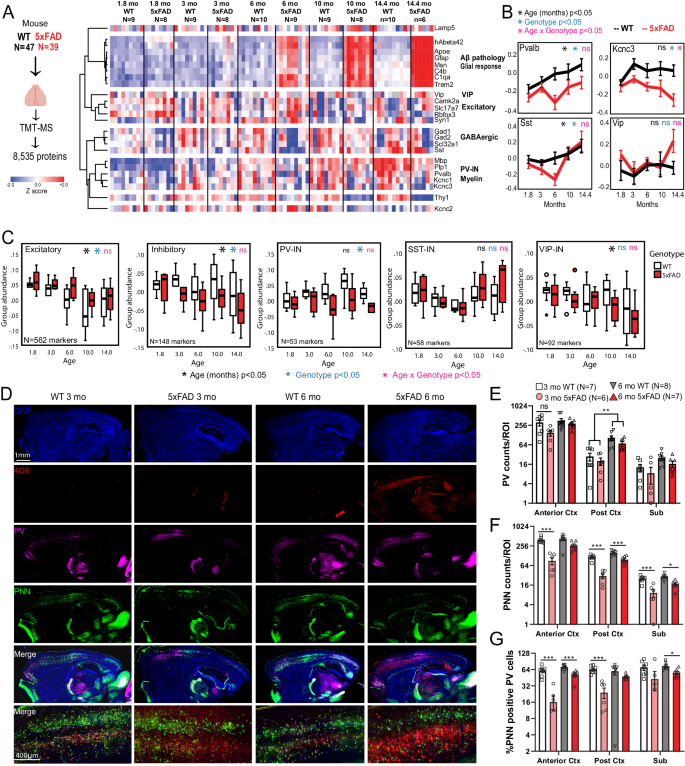
A Study outline for analysis of mouse bulk brain (cortex) TMT-MS proteomics data. 8,535 proteins were quantified by TMT-MS from 47 WT and 39 5xFAD mouse brains. From these, selected proteins reflective of Aβ pathology (hAβ42 peptide), were visualized as a heatmap after hierarchical clustering based on protein IDs (N/group for panels AâC are indicated). B Trajectories of change in levels of PV-IN proteins (Pvalb, Kcnc3), SST-IN (Sst) and VIP-IN (Vip) based on age and genotype. Error bars represent SEM. Statistical tests included linear regression analyses with age, genotype and âage x genotypeâ interaction terms as covariates. Levels of significance of each are indicated. C Trajectories of change in overall levels of pan-excitatory, pan-inhibitory, as well as PV-IN, SST-IN, and VIP-IN proteins, based on age and genotype. We used lists of transcriptomic markers of these classes of neurons (from sc/snRNAseq datasets) that were at least 4-fold enriched in the class of interest over all other neuronal types. After normalizing and z-transforming proteomic data, neuronal class-based group abundance scores were calculated and compared across ages and genotypes. Linear regression analyses were performed using age, genotype and age x genotype interaction term as covariates. Levels of significance of each, are indicated (Box plots: Median, inter-quartile range, min and max are shown). D Representative images from immunofluorescence microscopy studies of mouse brain (sagittal sections, WT and 5xFAD, ages 3 and 6 months, animals used for TMT-MS studies in A), to detect PV-INs (Pvalb protein), perineuronal nets (WFA lectin), Aβ pathology (4G8) and DAPI. 4x tiled images and 20x images from cortex are shown. EâG Quantitative analysis of Pvalb protein, PNNs, and proportion of Pvalb+ INs that have PNNs in the cortex and subiculum of WT and 5xFAD mice at 3- and 6- mo of age. Y-axes are log2 transformed. Error bars represent SEM (Post-hoc Tukey HSD pairwise comparisons; *pâ<â0.05, **pâ<â0.01, ***pâ<â0.005). See Supplementary Fig. 5 and Supplementary Data 4. Source data are provided as a Source Data file. Image was created using BioRender.com.
Based on this evidence of concordance of pathological changes across species based on our and recent proteomics studies73, despite evident inter-species differences between humans and mice, we focused our analysis to canonical transcriptomic markers (Supplementary Data 4) of different neuronal classes49, and identified distinct patterns of change of neuronal proteins with aging and genotype (5xFAD vs WT), as well as biological interaction between aging and genotype. Excitatory neuronal proteins (Camk2a, Slc17a7) showed an age-dependent decrease without an impact of genotype. In contrast, Pvalb and Sst proteins (found in PV-INs and SST GABAergic INs, respectively) increased with age in WT mice, but this trend was significantly blunted by 5xFAD genotype, particularly at 6 months of age (Fig. 4B). Kcnc3, which encodes a Kv3 channel highly expressed by PV-INs, showed an age-dependent decrease in 5xFAD mice while Vip, a marker of VIP-INs, did not show changes related to either age or genotype. We also observed a strong positive correlation between PV-IN proteins (Pvalb and Kcnc3) and myelin proteins (Plp1 and Mbp) (Fig. 4A) indicative of cell-cell interactions between PV-INs and oligodendrocytes74.
Using lists of markers enriched in pan-excitatory, pan-inhibitory neurons, as well as in sub-classes of PV-INs, SST-INs and VIP-INs49, we calculated composite cell-type-specific abundance scores of neuronal classes, to identify age and genotype (5xFAD vs. WT) effects in the bulk proteomic data (Fig. 4C). Pan-excitatory markers decreased with aging, and were minimally higher in abundance in 5xFAD brains as compared to WT, regardless of age. Pan-inhibitory markers also showed an age-dependent decrease in abundance which became more pronounced in 5xFAD mice. Among major IN classes, PV-IN markers showed a gradual increase with age in WT mice, although this was suppressed in 5xFAD brain (Fig. 4B). In contrast, SST-IN and VIP-IN markers were unaffected by genotype (Fig. 4C).
To determine whether PV-IN protein changes in 3-6 month-old 5xFAD mice are related to changes in PV-IN cell numbers or PV-IN proteins75, we performed IHC studies on an independent set of 3 and 6 month-old WT (nâ=â7-8) and 5xFAD (nâ=â7-8) brains, and assessed Pvalb protein levels along with detection of peri-neuronal nets (PNNs) by Wisteria floribunda agglutinin (WFA) in the cortex and subiculum (Fig. 4DâG). PNNs are known to disproportionately encapsulate PV-INs in the brain and are key regulators of PV-IN excitability76,77,78. The number of Pvalb protein-positive neurons in the cortex and subiculum were not impacted by genotype (5xFAD vs WT) at 3 or 6 months of age (Fig. 4D, E, Supplementary Fig. 9) although the proportion of PNN-positive PV-INs was significantly lower in 5xFAD mice (Fig. 4G).
Overall, our analyses of mouse brain proteomic data identify unique age and Aβ-dependent changes that appear to differentially impact neuronal sub-classes, where PV-INs may be selectively vulnerable in early stages of pathology in 5xFAD mice13. Our histological studies suggest that observed proteomic changes in PV-IN proteins in 5xFAD brain is not explained by changes in the absolute numbers of PV-INs79, although the health of PV-INs may be perturbed, as indicated by loss of PNNs around PV-INs in early stages of Aβ pathology and Aβ-induced changes in PV-IN proteins abundances. To better understand the molecular basis for differential vulnerability of PV-INs to AD pathology with spatiotemporal resolution, neuron class-specific native state proteomic investigations using CIBOP are warranted.
Unique molecular signatures of PV-IN proteome in early stages of Aβ pathology in 3 month-old 5xFAD mice
To identify molecular events specifically occurring in PV-INs because of early AD pathology, we applied the PV-CIBOP strategy to achieve PV-IN-specific TurboID expression, biotinylation and fluorescent labeling in WT and 5xFAD mice (Fig. 5A). Three weeks after RO AAV injection and 2 weeks of biotinylation, mice were euthanized at 3 months of age. This early stage of Aβ pathology (i.e., significant Aβ burden with minimal plaque formation restricted to the subiculum) was chosen to capture potentially-modifiable disease mechanisms. IHC confirmed PV-IN-specific biotinylation in the cortex of WT and 5xFAD PV-CIBOP mice (Fig. 5B). Flow cytometry of enzymatically-dissociated cortex7 confirmed equal efficiency of PV-IN targeting across all groups (Fig. 5C). Consistent with early stage of pathology, we observed minimal extracellular Aβ plaque pathology in the subiculum and cortex of 5xFAD mice, while total Aβ42 levels measured by ELISA were substantially increased in 3 month-old 5xFAD mice (Fig. 5D). WBs of cortical lysates showed robust biotinylation and V5 protein signals in all PV-CIBOP mice (Fig. 5E). WB of SA-enriched samples showed enrichment of biotinylated proteins in PV-CIBOP animals compared to controls, regardless of 5xFAD or WT genotype (Fig. 5F, Supplementary Fig. 10A).
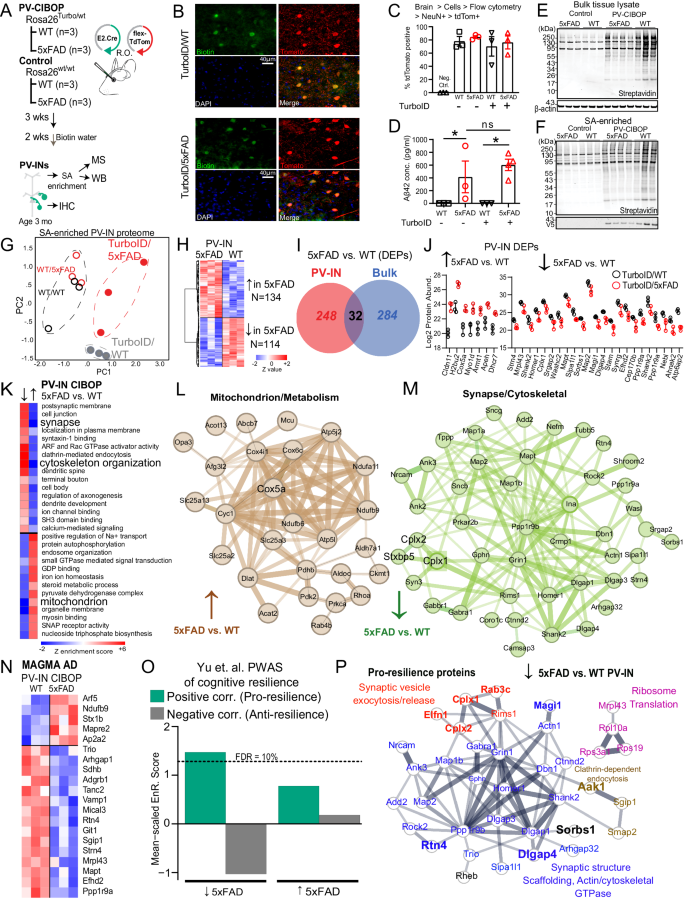
A Experimental outline for PV-CIBOP in 3-month-old 5xFAD mice. B IHC studies confirming PV-IN-specific biotinylation in WT and 5xFAD PV-CIBOP mice. C Flow cytometry analyses showing AAV-mediated targeting efficiency of PV-INs across experimental animals (nâ=â3/group; One-way ANOVA, pâ=â0.78; Data shown as meanâ±âSEM). D Aβ42 ELISA measurements from bulk cortex homogenates, confirming comparable Aβ42 levels across groups (nâ=â3 each for WT, FAD and Turbo WT, nâ=â4 Turbo 5xFAD, Data shown as meanâ±âSEM *pâ<â0.05, unpaired two-tailed T-test). E, F WB from bulk cortical tissue lysates and from SA-enriched samples showing robust biotinylation in PV-CIBOP compared to non-CIBOP mice. G PCA of MS data from SA-enriched proteomes: All PV-IN proteomes clustered away from control samples, and further distinction was observed between 5xFAD and WT PV-IN proteomes. H Heatmap representation of DEPs comparing WT/PV-CIBOP and 5xFAD/PV-CIBOP SA-enriched proteins. I PV-IN-specific DEPs minimally overlap with bulk tissue DEPs in 5xFAD and WT mice. J Top DEPs (showing at least 4-fold differential enrichment) comparing 5xFAD to WT PV-IN proteomes are shown (nâ=â3/group, meanâ±âSEM shown). K GSEA of DEPs comparing 5xFAD to WT PV-IN proteomes. L, M STRING protein-protein-interactions (PPI) within DEPs identified in Mitochondrial (L, increased in 5xFAD PV-IN) and Synaptic/Dendritic/Cytoskeletal (M, Decreased in 5xFAD PV-IN) ontologies. Thickness of edges indicates strength of known interactions. N Heatmap representation of DEPs comparing 5xFAD to WT PV-IN proteomes, limited to proteins encoded by genes with known genetic risk associations in AD (AD-MAGMA significance pâ<â0.05). O Enrichment of pro-resilience and anti-resilience proteins (from Yu et. al. PWAS study) within lists of DEPs (5xFAD vs. WT PV-IN proteomes). FDR 10% threshold is shown. P STRING PPIs of PWAS-nominated proteins positively associated with cognitive resilience (pro-resilience) that are decreased in 5xFAD PV-INs based on PV-CIBOP studies. Colors indicate shared functions and/or ontologies. Of these, proteins that are also selectively enriched in PV-INs as compared to Camk2a neurons (from CIBOP studies in Fig. 3) are highlighted (larger font, and bold). See Supplementary Fig. 6 and Supplementary Data 5 for related analyses. Source data provided as a Source Data file.
LFQ-MS of bulk brain samples (inputs) and SA-enriched (i.e., PV-specific) samples quantified 3,086 proteins and 2,149 proteins respectively (Supplementary Data 5). PCA of bulk proteomes indicated minimal effects of biotinylation on the overall brain proteome (Supplementary Fig. 10B). 1,973 proteins were enriched in the PV-IN proteome from both WT and 5xFAD PV-CIBOP groups as compared to negative control SA-enriched proteomes. Top PV-IN enriched proteins identified in WT/PV-CIBOP tissues in this experiment, including equal levels of enrichment of three Kv3 (3.1, 3.2, 3.3) channel proteins, agreed with findings in the first PV-CIBOP study in Fig. 1 (Supplementary Fig. 10C). PCA resolved differences between PV-CIBOP proteomes from WT and 5xFAD mice (Fig. 5G). We identified 248 differentially enriched proteins (DEPs) in PV-CIBOP proteomes from Turbo-expressing WT and 5xFAD mice, which included 134 proteins that were increased and 114 decreased in 5xFAD PV-INs (Fig. 5H, Supplementary Data 5). In alignment with our physiology studies33, Kv3 protein levels did not vary in young 5xFAD mice (pâ=â0.75, 0.10, 0.25 for Kv3.1, 3.2, and 3.3, respectively; unpaired T-tests). Overall, the PV-IN-specific DEPs in 5xFAD mice had very little overlap with DEPs identified in analyses of bulk brain proteomes from the same mice (Fig. 5I, Supplementary Data 5, Supplementary Fig. 10D). At the level of gene ontologies identified using gene set variation analyses (GSVA), the ontologies differentially enriched in the PV-IN proteome also showed minimal agreement with changes occurring in the bulk proteome (Supplementary Data 5). One example of stark discordance between bulk and PV-IN proteomic changes was the oxidative phosphorylation/aerobic respiration gene set, which was increased in PV-INs but decreased in the bulk proteome (5xFAD vs. WT). In contrast, synaptic proteins were decreased in both bulk and PV-IN proteomes, but the magnitude of this decrease was larger in PV-INs compared to bulk proteomes (Supplementary Data 5). This observed lack of concordance between the effects of 5xFAD genotype on the PV-IN proteome as compared to the bulk brain proteome, suggests unique metabolic/respiratory perturbations in PV-INs, that are most likely diluted in bulk tissue analyses.
Among DEPs identified in the PV-IN proteome, top proteins showing at least 4-fold increase in 5xFAD PV-INs included Cox5a, Dhcr7 and Apeh (Fig. 5J). Cox5a is a Complex IV mitochondrial protein involved in ATP synthesis80. Dhcr7 encodes 7-dehydrocholesterol reductase that catalyzes final rate limiting steps of cholesterol biosynthesis81. Apeh encodes acylaminoacyl-peptide hydrolase that hydrolyses terminal acetylated residues in small acetylated peptides, including degradation of monomeric and oligomeric Aβ82. Synaptic structural proteins including Shank2, Homer1, Map2 were conversely decreased by at least 4-fold in 5xFAD PV-INs (Fig. 5J). GSEA and GSVA identified in PV-IN proteomes showed that mitochondrial function, steroid biosynthesis, small GTPase signaling, and GDP binding were increased in 5xFAD PV-INs while structural/cytoskeletal, synaptic, axonal and dendritic ontologies were decreased in 5xFAD PV-INs (Fig. 5K, L, Supplementary Data 5).
We identified 36 post-synaptic proteins (post-synapse GO:0098794) that showed decreased levels in 5xFAD PV-INs, including structural constituents of the post-synapse (Dlgap1, Homer1, Gphn, Ina, Shank2, Git), enzymes with kinase activity or binding (Ppp1r9b, Bcr, Rtn4, Rheb, Prkar2b, Map2), neurotransmitter receptors (Grin1, Gabra1, Gabbr1), dendritic spine proteins (Grin1, Homer1, Dlgap3, Shank2, Dbn1, Bai1, Tanc2, Ncam1) and ribosomal subunits (Rps19, Rpl10a). Pre-synaptic proteins involved in synaptic vesicle fusion and exocytosis, including Complexins (Cplx1, 2), showed decreased levels in 5xFAD PV-INs (Fig. 5M). Of note, Cplx1 and 2 (but not Cplx3) were more also abundant in PV-INs as compared to Camk2a neurons in our CIBOP studies (Supplementary Fig. 10), in line with a recent proteomic study comparing excitatory and inhibitory synaptosomal fractions83. Several MAGMA-identified AD genetic risk factors also showed differential abundances in 5xFAD PV-INs as compared to WT PV-INs (e.g., decreased- Ppp1r9a, Mapt, Git1; increased- Arf5, Ndufb9, Stx1b (Fig. 5N).
To predict whether changes in the PV-IN synaptic protein landscape would result in detrimental or protective consequences, we cross-referenced the 5xFAD vs. WT PV-IN DEP list against pro-resilience and anti-resilience brain proteins identified in human studies (Supplementary Data 5)36. We found that pro-resilience proteins were over-represented while anti-resilience factors were under-represented in proteins that decreased in 5xFAD PV-INs (Fig. 5O). The pro-resilience proteins which were decreased specifically in PV-INs in early 5xFAD pathology included synaptic structural, synaptic scaffolding, actin/cytoskeleton (Dlgap13/4, Shank2, Homer1, Dbn1, Map1b, Map2, Ank3), ribosome (Rpl10a, Rps19, Mrpl43), mTOR-C1 regulating protein (Rheb), clathrin-dependent endocytic (Aak1, Sgip1, Smap2) and synaptic vesicle fusion/exocytosis/release related proteins (Cplx1, Cplx2, Elfn1, Rab3c, Rims1) (Fig. 5P). Many of these pro-resilience proteins that were decreased in 5xFAD PV-INs (e.g. Cplx1, Cplx2, Elfn1, Rab3c, Rtn4, Dlgap4, Sorbs1, Magi1) were also highly-enriched in PV-INs as compared to Camk2a neurons (Fig. 5P, Supplementary Fig. 10E, F), indicating that the changes in early AD pathology may indeed be related to PV-IN dysfunction.
Overall, these mouse-human integrative analyses indicate that altered levels of PV-IN bouton and dendritic proteins at early stages of Aβ pathology may result in detrimental synaptic or network effects, and thus represent proteomic changes potentially related to cognitive resilience with AD pathology. Thus, we next aimed to understand the functional impact of these PV-specific synaptic proteomic alterations.
Early Aβ pathology impacts PV-pyramidal cell neurotransmission and network activity
We leveraged the âE2â enhancer to express optogenetic actuators in PV-INs (Fig. 6, Supplementary Fig. 11) to measure PV-specific neurotransmission properties, where postsynaptic pyramidal cell recordings represent an integrated response to PV-specific, action potential-evoked neurotransmission. C1V1 (AAV.E2.C1V1) was injected in both WT and 5xFAD mice simultaneously with AAV.Camk2.YFP, which served to confirm accurate viral targeting (Fig. 6A). Acute slices were taken from two separate age cohorts (â~â2 or ~3 months old, 7 or 14 weeks respectively) and voltage clamp recordings were obtained from postsynaptic L5 pyramidal neurons (Fig. 6B). Short (â~â0.2âms) LED-light pulses (590ânm) reliably evoked IPSCs every trial (0.1âHz inter-trial interval) in both WT and 5xFAD recordings with minimal temporal jitter. The amplitude of the first IPSC amplitude was unchanged in both 2- and 3-month-old 5xFAD mice (e.g., 2-month-old WT: 133.5â±â25.90 pA, nâ=â10; 3-month-old 5xFAD: 163.7â±â30.55 pA, nâ=â7; pâ=â0.46, unpaired t-test), suggesting that GABAA receptor availability was similar in the postsynaptic pyramidal cells. As several synaptic vesicle fusion/exocytosis/release related proteins were altered in our 5xFAD PV-IN proteome, we more closely evaluated presynaptic properties (i.e., release probability) of PV-pyramidal synapses.
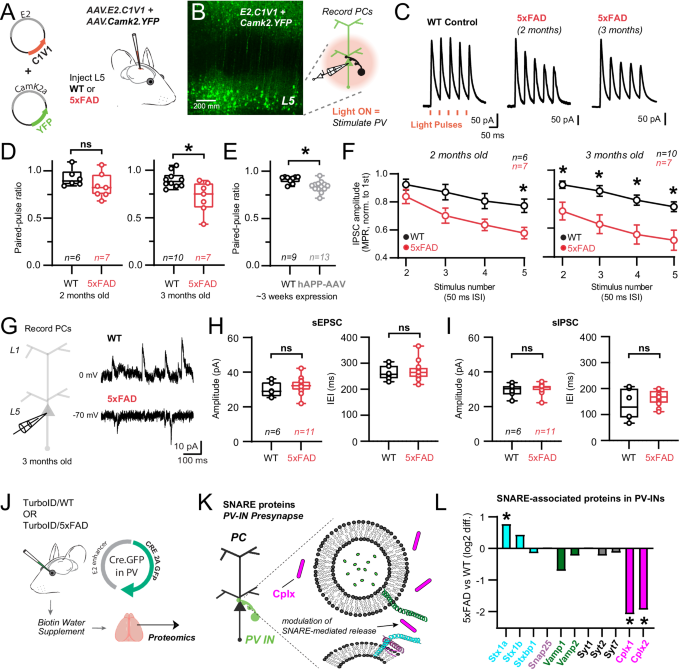
A E2.AAV-strategy for optogenetic activation of S1 PV-INs in WT and 5xFAD mice and co-labeling of Camk2a pyramidal neurons. B 2-photon image showing successful targeting of L5 following ~1 week after stereotactic surgery. Cartoon: Experimental workflow to stimulate PV-INs and record their synaptic properties in post-synaptic pyramidal cells (nâ=â3 mice/condition). C Averaged voltage-clamp traces from postsynaptic WT and 5xFAD pyramidal cells in layer 5 in response to short-amber light pulses. PV-IN IPSCs shown as time-locked to amber light pulses in a 20âHz train. D Quantification of paired-pulse ratios (PPR) of C1V1-evoked IPSCs in pyramidal cell recordings for 2- and 3-month-old WT and 5xFAD mice (2 months: pâ=â0.21; 3 months: pâ=â0.008). E Quantification of PPR of AAV1.DIO.ChETA-evoked IPSCs in pyramidal cell recordings from PV-Cre mice~3 weeks following hAPP-AAV injections in L5 (*pâ=â0.001). F Quantification of changes in PV synapse dynamics (multiple pulse ratio/MPR) in WT and 5xFAD mice (2 months: (left to right) pâ=â0.63, pâ=â0.07, pâ=â0.07, *pâ=â0.03; 3 months: (left to right) *pâ=â0.04, *pâ=â0.005, *pâ=â0.003, *pâ=â0.004; Two-way ANOVA with Sidakâs posthoc comparisons for each stimulus in WT and 5xFAD experiments). Data are displayed as meanâ+â/- SEM. G Voltage-clamp experiments were performed examining spontaneous synaptic activity in 3-month-old WT and 5xFAD mice. Holding voltage was interleaved between -70 and 0âmV to resolve spontaneous EPSCs and IPSCs, respectively. H Quantification of spontaneous EPSC amplitude and frequency in 3-month-old 5xFAD and WT mice. Data points indicate average from all spontaneous events from individual recordings (sEPSC Amplitude: pâ=â0.36); (sEPSC IEI: pâ=â0.69). I Quantification of spontaneous IPSC amplitude and frequency in the same recordings as in (H). Data points indicate an average value from all spontaneous events from individual recordings (sIPSC Amplitude: pâ=â0.67); (sIPSC IEI: pâ=â0.25). For box plots D-I: Median, inter-quartile range, min and max are shown. JâL Pre-synaptic PV-IN SNARE proteins identified by CIBOP, that show differential enrichment in 5xFAD vs. WT mice (Average log2FC enrichment is shown, *pâ<â0.05). Unpaired two-tailed t-tests used for all comparisons, except panel F. See Supplementary Fig. 7 and Source data files. Image was created using BioRender.com.
To examine whether modification of vesicle fusion and association proteins affected release probability and presynaptic dynamics at PV-pyramidal synapses, we measured the paired pulse ratio (PPR)84 and multiple pulse ratio (MPR) of optogenetically-evoked IPSCs at 20âHz using 1.5âmM external Ca2+ (Fig. 6C)85,86. Evaluation of the paired-pulse ratio showed modest depression (PPRâ~â0.9) in WT mice at both age timepoints. In 2-month-old 5xFAD mice, no difference in PPR was observed (Fig. 6D) although synaptic depression did intensify in 5xFAD mice during repetitive stimuli (MPR) at this age (Fig. 6F). However, in 3-month-old 5xFAD mice, a decrease in PPR emerged (Fig. 6D) and this more robust depression was now maintained throughout the stimulus train (Fig. 6F). Together these results show progressive presynaptic dysfunction in PV-pyramidal synapses following early Aβ pathology, consistent with changes in proteins regulating vesicular release probability (Fig. 5). Other mechanisms, such as proteins involved with vesicular docking and replenishment, axonal action potential signaling, and Ca2+ dynamics could also contribute. For example, changes in presynaptic parvalbumin expression may affect short-term plasticity and release probability87. However, we did not observe a change in bulk parvalbumin protein by MS (Fig. 4B) or IHC (Supplementary Fig. 9) at 3 months of age.
We next asked whether the signature of synaptic dysfunction observed in 5xFAD mice could be recapitulated in an independent, adult-onset model of APP/Aβ pathology. We packaged the human APP gene (variant NM_000484.4) into an AAV (AAV.Ef1a.hAPP). This particular APP isoform was chosen as it has been shown to proportionally increase with aging and is associated with increased AD risk88,89. While overexpression of hAPP is expected to produce a significant increase in Aβ90, this approach is distinct in that APP/Ab production is limited to the mature circuit, as in sporadic AD. 5-11 week old PV-Cre mice were co-injected with hAPP-AAV and AAV.DIO.CAG.ChETA for PV-specific optogenetic control Supplementary Fig. 11A. Control mice only received AAV.DIO.CAG.ChETA with saline to normalize overall viral titer across experiments. After 2-3 weeks expression, voltage clamp recordings were obtained from postsynaptic pyramidal neurons using 1.5âmM external Ca2+. Brief (~4âms) LED-light pulses (470ânm) could reliably evoke IPSCs on every trial (0.1âHz inter-trial interval) in both control and hAPP-AAV groups, with minimal temporal jitter Supplementary Fig. 11B,C. Similar to 3-month-old 5xFAD mice, the amplitude of the first PV-PC IPSC was unchanged after hAPP expression (Control: 68.18â±â38.37 pA, nâ=â9; hAPP-AAV: 72.70â±â44.49 pA, nâ=â13; pâ=â0.82, unpaired t-test). Optogenetic stimulation with ChETA also showed modest PV-PC synaptic depression in controls (Fig. 6D vs 6E). While PV-pyramidal synapses are often regarded as strongly depressing, their PPR is often observed as only slightly depressing when external Ca2+ is set at naturalistic levels91,92,93,94, as we found here. Indeed, after elevation of external Ca2+, we found a far more strongly depressing PPR (Supplementary Fig. 11EâG). Furthermore, basal synaptic properties were not attributable to direct Ca2+ entry through the rhodopsin pore locally at the presynapse (Supplementary Fig. 11G). Overall, similar to experiments in 5xFAD, synaptic depression measured via PPR and MPR was again enhanced following adult-onset hAPP-AAV expression (Fig. 6E; Supplementary Fig. 11D). Together these results complement our findings in 5xFAD mice, highlighting the early emergence of presynaptic dysfunction at PV-pyramidal synapses following early Aβ pathology.
Alterations in evoked release in PV-INs may affect basal network excitability by disrupting excitatory/inhibitory balance. Thus, we next examined whether changes in the amplitude and frequency of spontaneous EPSCs and IPSCs were apparent in pyramidal cell recordings from 3 month old 5xFAD mice (Fig. 6G). Interestingly, no changes were observed in either the amplitude or frequency of excitatory and inhibitory spontaneous synaptic events (Fig. 6 H, I) echoing recent work from 3-month-old 5xFAD mice in hippocampal slices, where local circuit behavior and oscillations were also largely resilient to change95. This may be due to several factors, including the relatively low AP frequency of evoked PV firing in the slice. APP/Aβ pathology could also preferentially affect evoked release, but spare stochastic vesicular fusion, as these two processes may be mechanistically independent in the same synapse96.
Potential molecular mechanisms of APP/Aβ pathology on altered neurotransmission
Our optogenetic findings indicate a selective disruption in evoked release at PV-pyramidal synapses. Thus, we next examined relevant SNARE-associated protein changes in 5xFAD mice using PV-CIBOP (Fig. 6J, K). Despite the fact that CIBOP does not explicitly target or isolate synaptic proteins, such as with more formal synaptosomal fractionation approaches, we still identified major SNARE-associated proteins in our proteomic datasets in both WT and 5xFAD mice (Fig. 6L) including Syntaxins, Snap25, Vamp, Synaptotagmins, and Cplx. Among this group of PV-IN SNARE associated proteins, only Cplx1 and 2 were significantly reduced in 3 month old 5xFAD. Interestingly, previous work has found that the loss of Cplx1 and 2 can alter evoked release without affecting spontaneous release, analogous to our findings in 5xFAD above97.
Evidence for extensive mitochondrial protein changes in PV-INs in response to early Aβ pathology
The extensive cell-type-specific proteomic alterations we observed in PV-CIBOP and bulk proteomes thus may be reflective of early homeostatic responses to maintain overall circuit functionality98,99, or potentially related to metabolic stress100, either directly or indirectly relating to Aβ pathology. Therefore, we next sought to explore the extent of changes to mitochondrial proteins and associated metabolic pathways specifically in PV-INs in early stages of Aβ pathology. Of 300 mitochondrial proteins (MitoCarta 3.0) biotinylated in PV-INs101, 30 proteins were increased (e.g. Cox5a, Mpst, Ndufa11, Ckmt1) and 4 proteins that were decreased (Mrpl43, Septin4, Sdhb, Bphl) in 5xFAD compared to WT PV-IN proteomes (Fig. 7A). Proteins involved in complex III, complex IV, complex V, amino acid metabolism and protein homeostasis were differentially enriched in 5xFAD PV-IN proteomes, while mitochondrial central dogma (mitochondria-specific DNA, RNA and translation-related elements), complex II and detoxification related proteins were unaffected (Fig. 7B). In contrast to the PV-IN proteome, only 26 mitochondrial proteins were differentially enriched in the bulk brain proteome, which included only 4 shared DEPs (Fig. 7C). Furthermore, the overall level of concordance between bulk brain and PV-IN mitochondrial protein levels was negligible (R2â=â0.0001). The increase of Cox5a levels in 5xFAD PV-INs but not in the bulk brain tissue was validated by Western Blot, using both bulk brain homogenates and PV-specific enriched proteins (Fig. 7D). In bulk brain MS proteomes, Cox5a showed age-dependent decrease in 5xFAD mice (Fig. 7E), a pattern in stark contrast to increased levels in PV-INs at 3 months. This overall pattern of increased abundances of mitochondrial proteins belonging to most mitochondrial compartments, may represent increased mitochondrial biogenesis to meet increased energy demands needed to sustain PV-IN functionality, particularly in the setting of emerging synaptic defects in early Aβ pathology.
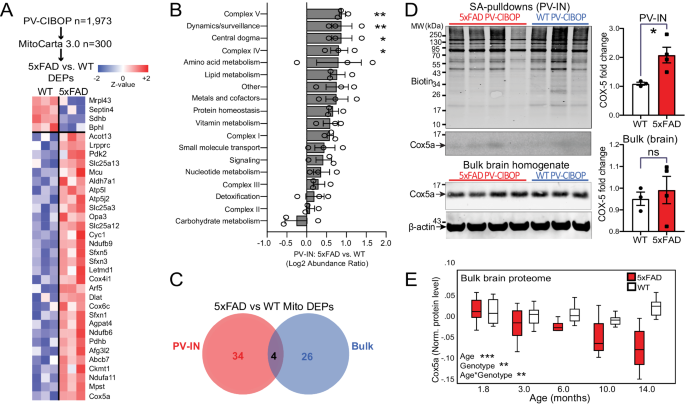
A Heatmap representation of mitochondria-localized proteins that were also identified as DEPs comparing 5xFAD to WT PV-IN proteomes. B Differential abundance analysis of distinct mitochondrial functional groups, comparing 5xFAD to WT PV-IN proteomes. The 300 mitochondrial proteins identified in PV-INs were categorized based on known functional and localization-related annotations (from MitoCarta 3.0). Protein levels were normalized and them group-wise abundances were estimated and compared across WT and 5xFAD genotypes (nâ=â3 (WT), 4 (5xFAD) *pâ<â0.05, **pâ<â0.01 for unpaired two-tailed T-test, error bars represent SEM). C. Venn diagram of mitochondrial proteins that were identified as DEPs in either PV-IN proteomes or bulk brain cortical proteomes, comparing 5xFAD and WT mice. Minimal overlap in DEPs were observed, highlighting unique mitochondrial effects of Aβ pathology in PV-INs, not visible at the bulk tissue level. D WB verification of increased Cox5a protein levels in PV-INs in 5xFAD as compared to WT mice. SA-enriched pulldowns were independently performed from samples used for LFQ-MS studies. Cox5a protein band intensity was normalized to total biotinylation signal in the SA-enriched pulldowns, and to beta-actin in the bulk brain homogenates, and then compared across genotype (5xFAD vs. WT) (nâ=â3 (WT), 4 (5xFAD); Data shown as meanâ±âSEM; *pâ<â0.05, unpaired two-tailed T-test). E Cox5a protein levels, quantified by TMT-MS, from an independent set of cortical brain homogenates obtained from WT and 5xFAD mice (from Fig. 4). Using linear regression modeling, age, genotype and age x genotype interaction terms were tested for associations with Cox5a protein levels. As compared to WT brain where Cox5a levels were relatively constant with aging, Cox5a levels in 5xFAD brain showed age-dependent decrease after 6 months of age. This pattern was discordant with increased Cox5a in PV-INs in 5xFAD mice at 3 months. *pâ<â0.05, **pâ<â0.01, ***pâ<â0.005). Source data are provided as a Source Data file.
Evidence for decreased mTOR-C1 signaling in PV-INs in early Aβ pathology
The mitochondrial and synaptic derangements occurring in PV-INs in 5xFAD mice suggest that upstream signaling pathways may be dysregulated in PV-INs. Metabolic signaling pathways, including Akt/mTOR, are important regulators of mitochondrial biogenesis and turnover, as well as synaptic function in neurons while several MAPKs (ERK, p38 MAPK, Jnk) impact cell proliferation, synaptic function and survival102,103,104,105. We found that PV-CIBOP labeled 75 proteins involved in Akt/mTOR (eg. Mtor, Rptor, Eif4b) and MAPK (eg. Map2k1, Ras proteins, Pak2, Akt3, Mapk3, Mapk10) signaling pathways (Fig. 8A). Of these, few proteins showed increased (Rhoa, Prkca, Hras, Cacna2d1) and decreased levels (Eif4b, Mapt, Rheb) in 5xFAD PV-INs (Fig. 8B). Interestingly, these differential effects of Aβ pathology were only observed in PV-IN proteomes, and not in the bulk brain proteome with the exception of Rheb, highlighting the specificity of these alterations in PV-INs in early AD pathology (Fig. 8C).
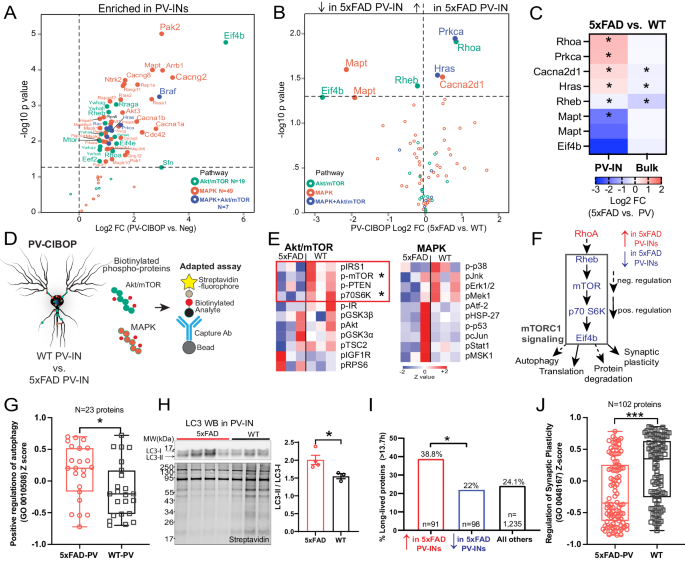
A. Akt/mTOR and/or MAPK signaling proteins biotinylated in PV-IN CIBOP proteomes (as compared to non-CIBOP mice). B. Akt/mTOR and MAPK proteins identified as DEPs comparing 5xFAD to WT PV-IN proteomes. C. Heatmap representation of Akt/mTOR and MAPK DEPs in PV-IN proteomes and their corresponding bulk brain proteomes (*pâ<â0.05, two-tailed unpaired T-test). D. Cartoon representation of adapted Luminex immunoassay to measure levels of PV-IN-derived phospho-proteins belonging to Akt/mTOR and MAPK proteins from bulk tissue. E. Heatmap visualization of Akt/mTOR and MAPK phospho-proteins in PV-INs measured by adapted Luminex assay from WT and 5xFAD mice (nâ=â3 mice/group, pâ<â0.05 unpaired two-tailed T-test). F. Summary: Decreased activity in mTOR signaling in 5xFAD PV-INs as compared to WT PV-INs, based on total protein levels estimated by PV-CIBOP MS, and phospho-protein levels by the adapted Luminex approaches. G. Comparison of proteins that positively regulate autophagy (GO:0010508), in 5xFAD and WT PV-IN proteomes (protein levels were normalized, z-transformed and then group-averaged across biological replicates before group comparisons; unpaired two-tailed T-test,*pâ<â0.05). H. Top: WB of PV-IN (SA-enriched) samples from 5xFAD and WT PV-CIBOP brain. LC3-II/I ratio was compared across the two groups. Bottom: Biotinylated protein from samples corresponding to WB images above. Data are displayed as mean values +/- SEM. (nâ=â3 (WT), 4 (5xFAD), *pâ<â0.05, independent two-tailed T-test). I. Analysis of DEPs (5xFAD vs. WT PV-IN proteomes) based on published protein half-lives in mouse brain. Proteins with increased levels in 5xFAD PV-INs were skewed towards proteins with longer half-lives (â>â13.7 days which represents the 75th percentile of protein half-lives in brain). This pattern is consistent with decreased translational efficiency and/or increased protein degradation, which would disproportionately impact the relative abundances of short-lived proteins. J. Comparison of proteins that regulate synaptic plasticity (GO:0048167) as a group, in 5xFAD and WT PV-IN proteomes (levels of 102 proteins were normalized, z-transformed and then averaged across biological replicates before group comparisons using unpaired two-tailed T-test (***pâ<â0.005). See Supplementary Data 6 for related analyses. Source data are provided as a Source Data file.
Based on the ability of CIBOP to biotinylate signaling proteins in PV-INs, we performed adapted Luminex assays to detect MAPK (Erk, P38 Mapk and Jnk) and Akt/mTOR signaling phospho-proteins, specifically derived from PV neurons8. In this approach, the biotinylated phospho-protein is immobilized on beads using capture antibodies, and then their biotinylation status is detected by streptavidin-fluorophore, to directly measure PV-IN-derived phospho-proteins from brain homogenates (Fig. 8D, Supplementary Data 5). We found that mTOR signaling (via phosphorylation of mTOR and down-stream target p70 S6K), was decreased in 5xFAD PV-INs while MAPK pathway activation was not altered (Fig. 8E). This pattern of decreased mTOR signaling was consistent with lower levels of Rheb (a direct activator of mTOR-C1 function), higher levels of RhoA (a known inhibitor of Rheb function) and lower levels of Eif4b (involved in translation initiation) in 5xFAD PV-INs (Fig. 8F)106. Collectively, our MS PV-IN CIBOP and adapted Luminex analyses indicate decreased mTOR-C1 activity in 5xFAD PV-INs.
To assess functional relevance of decreased mTOR-C1 in 5xFAD PV-INs, we assessed three composite measures of mTOR-C1 signaling, including autophagy (mTOR-C1 inhibits autophagy)105, translational efficiency/protein degradation (mTOR-C1 increases translational efficiency and decreases protein degradation)107,108 and synaptic plasticity (mTOR-C1 facilitates synaptic plasticity) (Fig. 8F)104. 23 GO-annotated positive regulators of autophagy (GO-0010508) were labeled in PV-INs and collectively, this group showed increased levels in 5xFAD PV-INs (Fig. 8G). Western blot analyses of biotinylated proteins from WT and 5xFAD PV-CIBOP mice found increased LC3-II (relative to LC3-I) in 5xFAD PV-INs as compared to WT PV-INs, consistent with increased autophagy in 5xFAD PV-INs (Fig. 8H). To determine whether decreased mTOR-C1 signaling in 5xFAD PV-INs impacts translational efficiency and/or increased protein degradation, we assessed the relative abundances of long-lived and short-lived proteins in 5xFAD and WT PV-IN proteomes. Using a reference dataset of protein half-life estimates derived from in vivo isotopic labeling studies in adult mice48, we found that DEPs that were increased in 5xFAD PV-INs were biased towards longer-lived proteins, as compared to non-DEPs as well as compared to DEPs with decreased levels in 5xFAD PV-INs (Fig. 8I, Supplementary Data 6). This molecular footprint of relatively-increased abundance of longer-lived proteins in 5xFAD PV-INs is consistent with decreased translational efficiency and/or increased protein degradation in 5xFAD PV-INs. Lastly, we found that proteins involved in regulation of synaptic plasticity (GO-0048167, nâ=â102 proteins) were also decreased in 5xFAD PV-IN proteomes (Fig. 8J), consistent with observed synaptic protein changes and physiological defects presented earlier (Figs. 5,6). These analyses provide congruent lines of evidence for decreased mTOR signaling at various levels of the signaling axis (upstream and downstream of mTOR-C1 function), that are associated with increased autophagic flux, decreased translational efficiency and decreased synaptic plasticity in PV-INs at early stages of Aβ pathology.