Collection and acclimatization under laboratory conditions
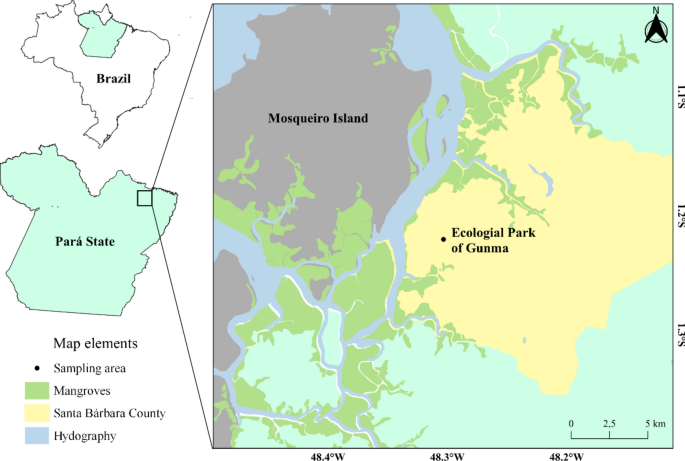
P. ephippifer is a terrestrial anuran species that reproduces mainly during the rainy season and is found in temporary pools at forest edges35, in addition, around 410 eggs are deposited in foam nests in the vegetation of ponds36, making them easy to detect during collection. Thus, three nests were collected in different temporary pools in forest edge areas in the Gunma Ecological Park located in the municipality of Santa Bárbara, Pará (Fig. 1, 01º13′00.86”S and 48º17′41.18”W) in the early morning hours of the same day.
Animal collection site, Gunma Ecological Park, Santa Bárbara—PA.
The nests were kept in containers with water from the puddles to prevent the eggs from drying out and transported to the laboratory. The nests were acclimatized for two days until hatching in the same container, with the aim of mixing larvae from different nests to maintain variability during the study. Acclimatization was carried out in plastic containers (36.5 × 23.5 × 7 cm) with a volume of 6 L, containing filtered water, constant aeration, in which the temperature (28 °C), pH (7.0), photoperiod (14/10 h) were controlled throughout the study. After hatching, the larvae were separated into four plastic containers under the same acclimatization conditions to avoid a high number of individuals in each container (120 individuals), and at each stage, larvae were collected randomly between containers. The animal collections were approved by the Biodiversity Authorization and Information System under protocol number 53433-1.
Experimental design
The tadpoles were fed once a day with commercial fish food (Novo Bits JBL—43% protein, 6% fat, 4% ether extract, 4% crude fiber, 10% mineral salts, 1.5% calcium) (0.250 mg), whereby food accumulation was avoided. The water was changed every second day, with removal of feces and possible food residues.
Larvae in three developmental stages were selected for the experiment: in premetamorphosis individuals between 28 and 29 Gosner stage (GS), in prometamorphosis between 38 and 39 GS and for the metamorphic climax all stages from 42 to 46.
The stages were determined on the basis of morphological parameters according to the Gosner table34 using a binocular stereomicroscope (model NSZ-606). The animals were then cryoanesthetized. No chemical reagent was used for anesthesia to avoid interference in oxidative stress parameters that are the focus of the study. All procedures were performed in accordance with the Brazilian legislation on the scientific use of animals (Law No. 11.794; October 8, 2008) and approved by the Ethics Committee for the Use of Animals (CEUA) of the Federal University of Pará, under the protocol 6371061016. All methods are reported in accordance with ARRIVE guidelines. The weight of the tadpoles was determined using an analytical balance (g), and the individuals were subsequently stored in 1.5 ml eppendorfs in an ultra-freezer at − 80 °C for further biochemical analysis.
The number of tadpoles collected varied at different stages according to the weight of the samples in order to quantify total proteins and to standardize and dosage of biochemical biomarkers. In the premetamorphosis, samples were pooled to achieve the required weight, with a total of 10 replicates of 20 larvae each. In all subsequent stages, samples were formed from only one individual. In prometamorphosis, 25 individuals were collected, and in metamorphic climax a total of 110 individuals were collected, covering stages 42–46.
Analysis of the biochemical biomarkers
Preparation of the samples
Samples were homogenized at a ratio of 1/4 (weight/volume) with a homogenization buffer and then centrifuged at 20,000 × for 20 min at 4 °C, according to Bainy et al.37. The obtained supernatant was aliquoted and stored at − 80 °C for later analysis.
Quantification of total proteins
Total proteins were analyzed using a commercial kit (Doles Ltda, Brazil) based on the Biuret assay (0.114 M trisodium citrate, 0.21 M sodium carbonate and 0.01 M copper sulfate). Measurements were performed in a multimodal microplate reader (Victor X3, Perkin Elmer) at 550 nm. The results are expressed in milligrams of protein/mL.
Glutathione S-transferase (GST)
Glutathione S-transferase activity was determined according to the method described by Habig, et al.38. In the analysis, the formation of the conjugate of 1-chloride-2,4-dinitrobenzene (CDNB) with reduced glutathione (GSH) catalyzed by Glutathione S-transferase in the sample is monitored for 1 min at 340 nm. Readings were performed in a spectrofluorimeter (Victor X3, Perkin Elmer) with a microplate reader. The results are expressed in UGST/ mg of protein, which corresponds to the amount of enzyme required to conjugate 1 μMol of CDNB/min/mg of protein at 25 °C and pH 7.0.
Total antioxidant capacity against peroxyl radicals (ACAP)
For the determination of ACAP, the method of Amado, et al.39 was used. This method is used to measure the antioxidant defense levels of organisms (enzymatic and non-enzymatic) exposed to radicals. It consists of the addition of ABAP (2′2′-azobis-2-methylpropiamidine dihydrochloride) in microplates together with biological samples and the fluorochrome H2DCF-DA (2′,7′-dichlorofluorescein). When heated to 37 °C, ABAP generates peroxyl radicals which are scavenged by the antioxidants present in the sample. If they are not scavenged, the radicals promote the oxidation of the fluorochrome, which then begins to fluoresce in the chemical form DCF (oxidized H2DCF-DA). Measurements were performed in a fluorescence microplate reader (Victor X3, Perkin 108 Elmer) for 30 min. Samples diluted to 0.5 mg of protein were used for all developmental stages. Results are expressed as the inverse of the relative area.
Lipid peroxidation (LPO)
The degree of lipid peroxidation was determined according to the protocol of Oakes and Van Der Kraak40, in which a by-product of lipid peroxidation, malondialdehyde (MDA), is quantified. The method analyzes the product formed in the reaction between MDA and thiobarbituric acid (TBA 0.8%) in an acidic environment (acetic acid, 20%) at high temperature (95 °C), forming MDA-TBA2. Butylatedhydroxytoluene (BHT) was used as an antioxidant in the samples, while 1,1,3,3-tetramethoxypropane (TMP) served as a standard. Sodium dodecyl sulfate (SDS, 8.1%) was used as a surfactant and n-butanol was used to separate the organic from the inorganic phase. Readings were performed in a fluorescence reader (515 and 553 nm for excitation and emission, respectively). The results were expressed in nM MDA/g of wet tissue, which represents the MDA lipid concentration (nM) in weight (g) of tissue.
Analysis of the gene-expression biomarkers
To evaluate transcriptional levels, the reverse transcription technique was performed followed by quantitative PCR (RT-qPCR). Total RNA was extracted using the PureLink RNA Mini Kit (Thermo Fisher Scientific, 12183018A) following the manufacturer’s instructions. RNA integrity (RIN–RNA Integrity Number) was assessed using the NanoVue spectrophotometer and 1% agarose gel. RNA was treated with DNase I (Thermo Fisher Scientific, EN0521) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized using the High Capacity kit (Thermo Fisher Scientific, 4368814) following the manufacturer’s instructions. PCR primers for genes related to oxidative stress (nrf2, gst, gsr, gclc) (Table 1). 1 µl of cDNA (30 ng/uL) was amplified using the RealQ Plus 2 × Master Mix Green, High ROX kit (Ampliqon, A323402) and 400 nM of each primer in a final volume of 20 uL. The cycling conditions were: 10 min at 95 °, 40 cycles of denaturation at 95 °C for 15 s and annealing and extension at 60 °C for 1 min. Expression levels were detected on the Bio-Rad CFX Maestro system. Actg gene transcriptional levels were used as a reference. The dissociation curve was evaluated to confirm specific amplification. Data were normalized using the Q-Gene program41,42.
Data analysis
A unidirectional analysis of covariance (ANCOVA) was performed to verify the influence of weight (covariate) on the response of the analyzed biomarkers (Glutathione S-transferase, Total antioxidant capacity and Lipid peroxidation). The independent variable corresponds to the stages of P. ephippifer and had three levels (premetamorphosis, prometamorphosis and metamorphic climax). The dependent variable was the results of the biomarkers expressed in each corresponding unit.
Initially, the assumption of homogeneity of the regression slope between the covariate and the variable was tested in all parameters. Subsequently, the normality of the data was tested using the Shapiro–Wilk test. If necessary, a Box-Cox transformation was applied to the data in order to meet normality assumptions. In addition, a Levene test was performed to check the homogeneity of the residual variances.
Differences in the responses of oxidative stress parameters (Glutathione S-transferase, Total antioxidant capacity and Lipid peroxidation) and genes expression were tested using a one-way analysis of variance (one-way ANOVA) followed by a post-hoc Tukey test. The data were analyzed in the program R, version 4.1.143 and for graphics with the package ggplot2.
For the comparison between the climax stages (42, 43, 44, 45, 46 GS), the assumptions of normality and homoscedasticity were tested using the Shapiro-Wilks and Levene tests. For Glutathione S-transferase and Lipid peroxidation data (parametric), a one-way ANOVA followed by a Tukey’s post-hoc test was used for pairwise comparisons of means. Results were expressed as mean ± standard error. The significance level was set as p < 0.05. Total antioxidant capacity (non-parametric), analyzes were performed using the Kruskal–Wallis test followed by the Nemenyi post-hoc test and expressed as median ± 1st quartile. The assumed significance level was 5% in all cases44.