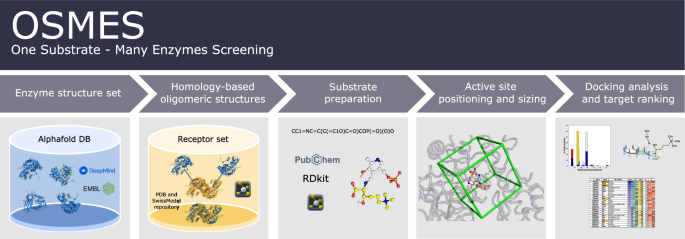
One substrate-many enzymes screening (OSMES) for PLP-dependent enzymes
Here we develop an automated procedure to perform a reverse docking screening of a substrate containing a primary amino group bound to PLP cofactor as a Schiff base (external aldimine; substrate), against a set of 3D enzyme structures of a selected PLPome (enzyme set) (Fig. 1).
Scheme of OSMES. The pipeline consists of 5 main steps performed automatically: (i) AlphaFold monomeric models for selected proteins are retrieved; (ii) oligomeric structures are determined with SWISS-MODEL templates; (iii) the substrate is prepared for docking and (iv) used to determine the gridbox size at the active site; (v) finally, the pipeline performs docking analysis and the results are ranked using different methods.
As an enzyme set we used PLPomes of Homo sapiens and Mus musculus retrieved from the B6 database (B6DB; http://bioinformatics.unipr.it/B6db) composed of 56 and 57 genes respectively. For each RefSeq accession number we obtained the corresponding UniProt ID to download the AlphaFold monomer28 and mark the position of the catalytic lysine useful for subsequent steps. In this first step, we discarded proteins without a conserved catalytic lysine, namely AZIN1, AZIN2, SPTLC1 and PDXDC1 in both sets and Ldc1 in the mouse set, obtaining 105 enzyme targets for our analysis (Supplementary Table 1). The vast majority of our targets have AlphaFold models of very high confidence (pLDDT>90 over 90% of residues) for the overall (>80%) and active site (>95%) residues (Supplementary Fig. 1).
Since most PLP-dependent enzymes belong to fold-type I, which is characterized by obligate dimeric association forming two identical active sites at the interface, oligomerization of the monomeric AlphaFold models is a crucial step of the OSMES pipeline. We therefore exploited models available in the SWISS-MODEL Repository (SMR; https://swissmodel.expasy.org/repository) as templates to assemble AlphaFold monomers into oligomeric structures (Fig. 1, step 2). In our set of enzymes, 96 structures were modeled as oligomers, mostly homomers (79 dimers, 13 tetramers) with the exception of SPTLC2 and SPTLC3, which were modeled as hetero-dimers, both associated with SPTLC1. Once the enzyme set is prepared, the procedure automatically builds the covalent adduct between PLP and a substrate molecule with a given PubChem ID, and creates a 3D coordinate file of the external aldimine for docking screening (Fig. 1, step 3). For each enzyme structure, the grid center for docking calculation is positioned at the NZ atom of the catalytic lysine, and the grid size is defined according to the size of the substrate (Fig. 1, step 4) (see Methods).
As a final step (Fig. 1, step 5) the pipeline runs the docking analysis of the substrate against each enzyme structure with AutoDock for Flexible Receptors (ADFR)35, choosing as flexible residue the same catalytic lysine used to place the grid. The results of the screening are then parsed to rank targets according to different methods (see below).
Evaluation of catalytically favorable conformations is the best performing metric in OSMES
Before proceeding with OSMES to our case study, we assessed the ability of different ranking methods to identify enzymes involved in particular PLP-dependent reactions. We considered 13 different substrates (Supplementary Fig. 2) against the two PLPomes (human and murine) for a total of 26 screenings evaluated with 7 ranking methods (Fig. 2). In each screening, one or more positive controls represented by enzymes known to catalyze the examined reaction (validation set) were considered. The validation set consisted of a total of 42 positive controls divided into 14 decarboxylases, 6 aldolases, 14 aminotransferases and 8 other reactions encompassing 4 ammonia-lyases, 2 γ-lyases, and 2 hydrolases (Supplementary Table 2). This set represents about 45% of the 93 human and mouse PLP enzymes with a four-digit EC number.
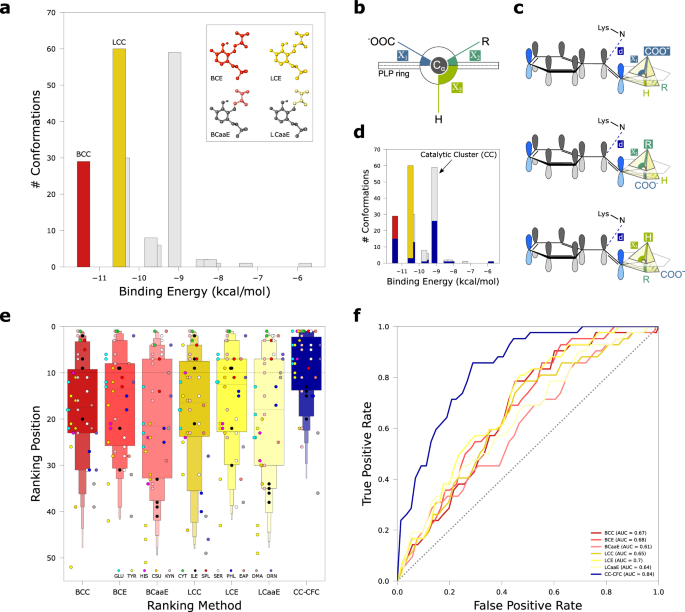
a Representation of the 6 ranking methods related to the best cluster (BC; red tones) and the largest cluster (LC; yellow tones). The bar plot represents the 200 conformations of a single docking run clustered with a 3âà RMSD threshold; LCC and BCC methods consider the number of conformations in the respective cluster. The atoms of the substrate considered in the energy-based ranking methods (BCE, LCE, BCaaE, LCaaE) are highlighted in the insets. b Scheme of the side view of the PLP pyridine ring and the three Cα bonds with the respective angles (Ï) with respect to the PLP ring plane. c Catalytically favorable conformations (CFC) in the three different PLP-dependent reactions. The conformations from docking analysis are considered CFC if the distance (d) between Nε of catalytic lysine and imine carbon is â¤5âà in the catalytic cluster, and the bond cleaved in the expected reaction (superior circumradius) is nearly orthogonal to the PLP ring (plane), that is its angle Ï has the maximum relative value (see Methods). d Bar plot highlighting in blue the number of CFC in different clusters. Black arrow indicates the Catalytic Cluster (CC) which does not always coincide with BC (red) or LC (yellow). e Letter-value plot showing the distribution of the validation set (nâ=â42) colored according to the 7 ranking methods. BC related methods are colored in red tones; LC related methods are colored in yellow tones; CC-CFC is colored in blue. Individual dots representing ranking position of positive controls (i.e., enzymes known to act on the substrate) are colored according to substrate (legend); black dashed line delimits the top 10 positions. The band indicates the median, the main box indicates the first and third quartiles with every further minor box splitting the remaining data into two halves. f Receiver operating characteristic curve (ROC) for the different ranking methods colored as in panel e; the dotted diagonal represents an area under curve (AUROC) value of 0.5. Source data are provided as a Source Data file.
Among the pose clusters obtained from ADFR analysis, we considered both the lowest-energy cluster (best cluster, BC) and the most populated cluster (largest cluster, LC) (Fig. 2a). The lowest-energy cluster, reflecting the stability of the system, is regarded as the energetically favored one. The most populated cluster, reflecting a higher conformational entropy of the system36, is regarded as the statistically favored one. For both BC and LC, we ranked the results using three different ranking methods: i) the number of conformations in the cluster (BCC and LCC); ii) the lowest binding energy of the cluster conformations (BCE and LCE); and, to discount the contribution on the constant moiety of the external aldimine, iii) the lowest binding energy of the cluster conformations without the PLP atoms, considering only the amino acid (BCaaE and LCaaE).
In addition to these more canonical criteria, we introduced a ranking method that evaluates the number of catalytically favorable conformations (CFC) based on Dunathanâs stereoelectronic hypothesis22. According to this widely accepted feature of PLP catalysis, when a compound containing a primary amine group binds covalently to the PLP cofactor to form the external aldimine, the reaction proceeds by breaking the bond more parallel to the Ï orbitals of the cofactor pyridine ring, or in other words, more orthogonal to the plane formed by the latter. In the case of an α-amino acid, three different cases are possible (Fig. 2b, c), represented by the breaking of the Cα-COOH (as in decarboxylases), Cα-Cβ (as in aldolases), and Cα-Hα bond (as in racemases, aminotransferases, and other lyases). On this basis, for every substrate in our screenings we considered CFC conformations in which the angle (Ï) with the PLP ring is maximum for the bond cleaved during the reaction (Ï1 for Cα-COOH; Ï2 for Cα-Cβ; Ï3 for Cα-Hα; Fig. 2b, c). As an additional condition for a CFC, we set an upper threshold of 5âà for the distance between the NZ atom of catalytic lysine and the imine carbon of external aldimine (Fig. 2c). The cluster with the maximum number of CFC is considered the âcatalytic clusterâ (CC) and scored by the number of CFC it contains (CC-CFC) (Fig. 2d).
The distribution of the validation test ranked with the 7 different ranking methods shows that with the CC-CFC method the positive controls are generally ranked higher than with other methods (Fig. 2e). Within the CC-CFC distribution, a difference in the performance emerged by categorizing positive controls according to the reaction type, with aminotransferases (A) achieving worse results with respect to other reactions (O) that break the Cα-Hα bond or aldolases (B) and decarboxylases (D) (Supplementary Fig. 3a, b). The good performance obtained by CC-CFC is supported by the area under the receiver operating characteristic (AUROC) that confirms the CC-CFC as the most performing ranking method, with an AUROCâ=â0.84 compared with 0.7 of LCE, the second best method (Fig. 2f). As an alternative for the second step of the OSMES pipeline (the assembly of oligomeric structures), we considered the use of AlphaFold Multimer37. Also in this case, CC-CFC was the best ranking method. However, a slight decrease in the performance was observed with respect to the use of oligomers based on SWISS-MODEL templates (AUROCâ=â082, Supplementary Fig. 4).
Application of OSMES to the identification of a missing gene in carnitine biosynthesis
Carnitine biosynthesis begins with release of N6-trimethyllysine (TML) from the breakdown of post-translationally modified proteins such as histones, calmodulin, cytochrome c, myosin, etc.38,39, and involves four enzymatic steps (Fig. 3a). Reactions 1 and 4 are catalyzed by two Fe2+-dependent dioxygenases: TML dioxygenase (TMLD) and γ-butyrobetaine dioxygenase (BBD), which are related by homology; reaction 3 is catalyzed by trimethylamino butyraldehyde dehydrogenase (TMABADH); reaction 2, the aldol cleavage of HTML to generate glycine and TMABA, is catalyzed by HTMLA. Although there is evidence that this activity requires PLP40,41, the molecular identity of HTMLA in mammals and other metazoans is unknown.
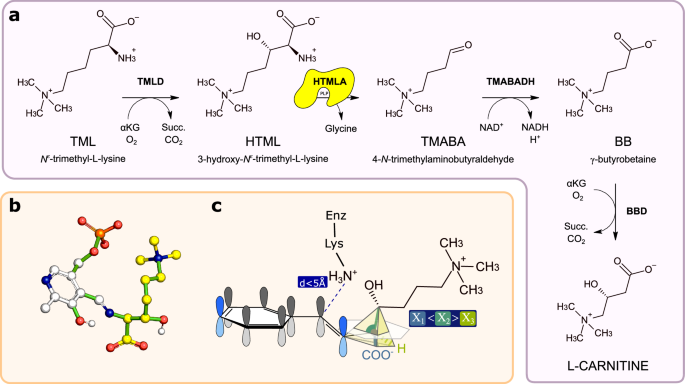
a Carnitine biosynthetic pathway in animals. HTMLA, the missing enzyme catalyzing the second step of the pathway is highlighted in yellow. b Atomic model of the energy-minimized conformation of the HTML-PLP external aldimine used for the OSMES procedure. In yellow the carbon atoms of HTML, in white the carbon atoms of PLP. Non-carbon atoms are colored according to CPK convention. Rotatable bonds are colored in green. c Expected geometry of the catalytically favorable conformation of the docked HTML-PLP substrate. The Cα-Cβ bond is considered labile when Ï1â<âÏ2â>âÏ3 and dââ¤â5âà . Source data are provided as a Source Data file.
The pathway described above is not universally present in eukaryotes. For instance, it lacks in the yeast Saccharomyces cerevisiae and the darkling beetle Tenebrio molitor, which require an external supply of carnitine for fat metabolism42,43. The distribution of the genes encoding TMLD and BBD in eukaryotes (Supplementary Fig. 5), shows that the known pathway for carnitine biosynthesis is especially present in opisthokonts (fungi and metazoa). However, absence of TMLD and/or BBD in several species, particularly in protostomes, suggests multiple pathway losses, a suitable condition for the identification of missing genes by coevolutionary analysis. This analysis, conducted with a sensitive method of gene coevolution44 in 1,952 eukaryotic genomes45 did not reveal an obvious HTMLA candidate, although the best signal among PLP-dependent enzymes was found for an orthogroup annotated as threonine aldolase (Supplementary Table 3). Interestingly, a gene belonging to this group has been previously implicated in Candida albicans as HTMLA30. A gene homologous to threonine aldolase (Tha1) is found in several mammals including mice, but not in humans46 nor in other species capable of synthesizing carnitine (Supplementary Fig. 5).
Since homology and coevolutionary analysis provided inconclusive evidence on the identification of mammalian HTMLA, we decided to use OSMES on the full set of PLP-dependent enzymes of human and mouse to identify candidates on a structural basis. To this end, we modeled the external aldimine PLP-HTML complex assuming free rotations around rotatable bonds (Fig. 3b) and defined the condition for catalytically favorable conformations of the docked substrate (Fig. 3c): a distance of â¤5âà of the PLP aldehyde carbon of the substrate from the NZ atom of catalytic lysine and a relative maximum for the Ï2 angle, as expected for the cleavage between Cα-Cβ that occurs in the HTMLA reaction (Fig. 3a).
HTMLA candidates revealed by OSMES in the human and mouse PLPome
The best performing method (CC-CFC) was used to rank the results of HTML-OSMES against human and murine PLPomes (Fig. 4a). In the two rankings orthologous enzymes are in similar positions, as confirmed by the correlation between the two sets (Spearman râ=â0.83; Supplementary Fig. 6).
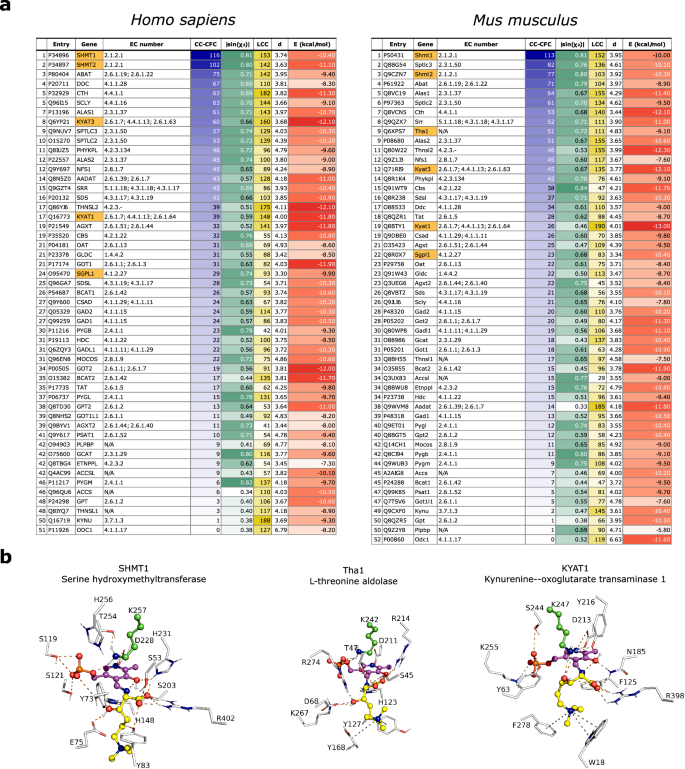
a HTML-OSMES against human and mouse PLPomes ranked with CC-CFC method. Best results (highest for LCC, CC-CFC, |sin(Ï2)|; lowest for E) in the columns are highlighted with darker colors. |sin(Ï2)|, d and E columns represent the mean values of CC. In orange are highlighted the enzymes with known β-lyase or aldolase activity. b Structural representation of the lowest-energy binding modes among the catalytic clusters obtained by docking of HTML-PLP substrate for SHMT1, Tha1 and KYAT1. Non-carbon atoms are colored according to CPK convention. The conformations are shown with ball-and-sticks and are composed of PLP cofactor (magenta) covalently bound to HTML (yellow), and flexible catalytic lysine (green). The binding site residues (â¤4.5âà from HTML-PLP) are shown in lines labeled with one-letter code and number. Polar interactions between substrate and protein are indicated with orange dashes, while cation-Ï interactions are indicated with olive dashes. Source data are provided as a Source Data file.
In both rankings, the first hit is the cytosolic serine hydroxymethyltransferase (SHMT1; Shmt1); its mitochondrial version (SHMT2; Shmt2) ranks just after in second (human) and third (mouse) position, as expected from the strong conservation of active sites residues (Supplementary Fig. 7). Interestingly, it has been shown that E.coli SHMT can act as an aldolase on β-hydroxylated amino acids, especially with erythro configuration47 that is the configuration adopted by HTML, and it has been proposed that SHMT could be responsible for HTMLA activity in mammals31. Descending with the ranking, other potential candidates with tested or predicted aldolase activity and belonging to the same KEGG Reaction Classes as HTMLA (RC00312 and RC00721) are found. These are sphingosine phosphate lyase (SGPL1, Sgpl1; EC: 4.1.2.27), an enzyme anchored to endoplasmic reticulum that catalyzes aldol cleavage forming phosphoethanolamine, and the putative mouse L-threonine aldolase (Tha1), not characterized experimentally but traceable by homology to the yeast low specificity L-threonine aldolase (GLY1, EC: 4.1.2.48). A GLY1 paralog has been genetically characterized as HTMLA in C. albicans30. Another example of promising candidates is the pair of paralogous enzymes called kynurenine aminotransferases (KYAT1, Kyat1, KYAT3, Kyat3). These enzymes catalyze the transamination of kynurenine into the corresponding α-keto acid. However, they are also able to catalyze β-lyase reactions toward cysteine-S-conjugate substrates (EC: 4.4.1.13), although the reaction mechanism involves deamination unlike HTMLA48.
In the catalytic clusters of all the mentioned candidates, ADFR is able to position the PLP cofactor in a binding mode similar to that observed in the available experimental structures of homologous enzymes in complex with PLP (Supplementary Fig. 8). In all four SHMTs and in Tha1, the lowest-energy conformations of HTML-PLP in the catalytic cluster have the Cα-Cβ bond more perpendicular than in the other enzymes (Fig. 4b, Supplementary Fig. 9). By contrast, in the case of both SGPL1 and Sgpl1 (Supplementary Fig. 9), and all KYATs (Fig. 4b, Supplementary Fig. 9), the Cα-COOH (Ï3â<âÏ1â>âÏ2) and Cα-Hα (Ï1â<âÏ3â>âÏ2) bond, respectively, are the most perpendicular and therefore in an unfavorable conformation for aldol cleavage.
In all four KYATs and Tha1, visual inspection of the docked complexes revealed the presence of an aromatic cage (Fig. 4b, Supplementary Fig. 9), characteristic of proteins that bind N-trimethylated substrates, establishing hydrophobic and cation-Ï interactions with the trimethyl ammonium group49. The constant presence of a quaternary amine group in the intermediates of carnitine biosynthesis (Fig. 3a), suggests that an aromatic cage could be a structural feature of all enzymes of the pathway, as evidenced by the BBD structure in complex with γ-butyrobetaine50, the conservation of the corresponding residues in its homolog TMLD, and the binding mode predicted by docking of the substrate in the TMABADH active site (Supplementary Fig. 10).
Biochemical validation of HTML-OSMES candidates
For the above reasons, screening candidates KYAT1, SGPL1, SHMT1 and SHMT2 from Homo sapiens, and Kyat3 and Tha1 from Mus musculus were chosen for the experimental validation. In addition, we considered screening candidates without previous evidence of aldolase or beta-lyase activity: human ABAT as an example of high-ranking hit, mouse Thnls2 and Oat as mid-ranking hits, and human PSAT1 as a low-ranking hit.
Each protein was produced using optimized conditions in recombinant form to be assayed for HTMLA activity. We obtained soluble expression for all the proteins with the exception of ABAT. Recombinant SHMT1, SHMT2, KYAT1, PSAT1, Kyat3, Thnsl2, Oat were obtained in pure and soluble form after overexpression in E. coli (Supplementary Fig. 11aâd; insets). In order to obtain recombinant SGPL1 and Tha1 in the soluble form (Supplementary Fig. 11e,f; insets), they were co-expressed with chaperones (GroEL/GroES) as truncated forms without the N-terminal membrane anchor and mitochondrial signal (Supplementary Fig. 12; see Methods). All the enzymes showed the typical spectrum of protein-bound pyridoxal phosphate in the ketoenamine tautomer, with a peak around 400â430ânm (Supplementary Fig. 11).
Stereospecific (2S,3S) HTML for the activity assays was obtained enzymatically from chemically-synthesized TML (see Methods) by exploiting the first reaction of the pathway (Supplementary Fig. 13). The activity assays show that SHMT1, SHMT2 and Tha1 catalyze the aldol cleavage of HTML; on the contrary, KYAT1, Kyat3, PSAT1, Thnsl2, Oat and SGPL1 are catalytically inactive towards HTML (Fig. 5).
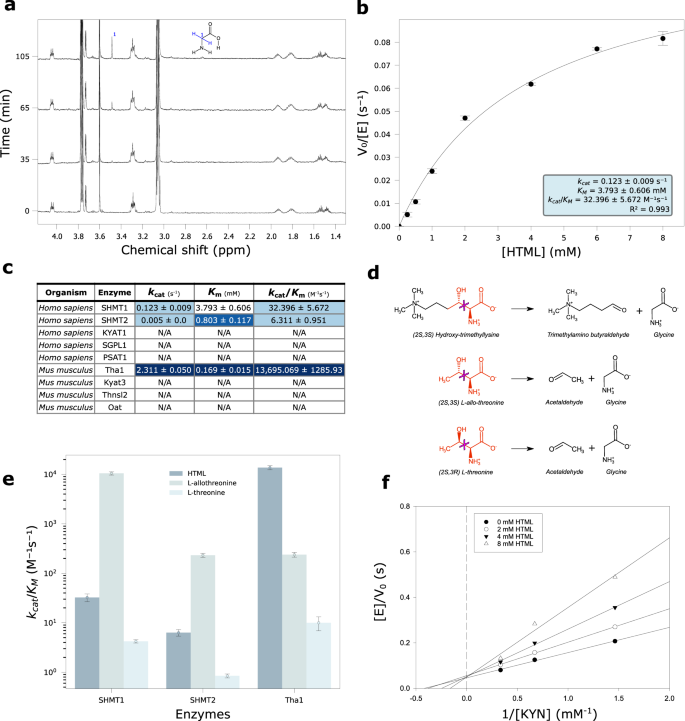
a Time-resolved 1H NMR spectra of SHMT1 activity in the presence of 5âmM HTML at 0, 35, 65 and 105âmin. Cα protons singlet of glycine is assigned in the structure. b Nonlinear fitting to the Michaelis Menten equation of the dependency on HTML concentrations of the initial reaction velocity of SHMT1 (1âμM). Data are presented as mean valuesâ±âSEM; nâ=â4 independent experiments for each point. c Kinetic parameters (kcat, Km, kcat/Km) of HTMLA reaction of tested enzymes with mean and standard deviation values obtained by nonlinear fitting. The blue-white gradient indicates better (blue) or worse (white) values in each column, where better means higher for kcat and kcat/Km or lower for Km. d Scheme of the broken bond (magenta cross) in the aldol cleavage reactions of HTML, L-allo-threonine, L-threonine. In red are the portions common to the three substrates. e Bar plot in log scale of the catalytic efficiency of different enzymes (Tha1, SHMT1, SHMT2) with different substrates (HTML, L-allo-threonine, L-threonine). The data points correspond to the kcat/Km values and the error bars represent the standard deviations of the fitting parameters. f Lineweaver-Burk double-reciprocal primary plot of the inhibition by HTML of kynurenine aminotransferase activity of KYAT1. The kynurenine concentration ranged from 0.75 to 3âmM. The concentrations of HTML were 0, 2, 4, and 8âmM. Source data are provided as a Source Data file.
Human SHMTs catalyze the aldol cleavage of HTML
In the 1H NMR spectrum of HTML after addition of SHMT1, the increase of a singlet at 3.55 ppm corresponding to glycine α-protons is visible (Fig. 5a), clearly appearing after 60âmin of reaction. TMABA formation is confirmed by 2 distinctive signals at 9.63 ppm and 5.05 ppm of the carbonyl proton and its hydrated form (geminal diol), respectively (Supplementary Fig. 14a).
Kinetic characterization of HTML cleavage catalyzed by SHMT1, carried out by a continuous spectrophotometric coupled assay that exploits NAD+ reduction signal at 340ânm in the presence of the third enzyme of the pathway (TMABADH), shows a dependence of the initial velocities on substrate concentrations following Michaelis-Menten kinetics (Fig. 5b). The fitting of data to the Michaelis-Menten equation reveals a catalytic efficiency (kcat/Km) of 32.17â±â5.34âsâ1 Mâ1 (Supplementary Table 4). We also characterized the enzymatic activity of SHMT2 by spectrophotometric assay (Supplementary Fig. 15i), and measured a lower catalytic efficiency (6.23â±â1.26âsâ1 Mâ1) compared to SHMT1 (Fig. 5c). In fact, despite a lower Km (0.80â±â0.16âmM vs 3.79â±â0.44âmM) SHMT2 is penalized by a worse kcat (0.005â±â0.000âs–¹ vs 0.122â±â0.006âsâ1). The aldolase activity of human SHMTs towards HTML was not affected by the presence of tetrahydrofolate, a cofactor in the hydroxymethyltransferase reaction catalyzed by the enzyme (Supplementary Fig. 16).
Mouse threonine aldolase (Tha1) shows higher HTMLA activity than human SHMTs
The 1H NMR spectrum of HTML after the addition of Tha1, shows peaks with the same chemical shift observed in the reaction with SHMT1, but in higher quantities (Supplementary Fig. 14b), suggesting the same enzymatic activity, but a different efficiency for the two enzymes. A small upfield shift is visible in the main peak of the trimethylated ammonium protons at 3.11 ppm (Supplementary Fig. 14b).
Kinetic characterization of Tha1 by the same spectrophotometric assay as SHMT1, and fitting to the Michaelis-Menten equation (Supplementary Fig. 15j) resulted in a kcat of 2.311â±â0.029âsâ¹ and Km of 0.169â±â0.009âmM. Comparison with SHMT1 shows better values for both Tha1 constants and a kcat/Km (1.36 Ã104âsâ1 Mâ1) about a thousand times greater (Fig. 5c). To test the substrate specificity of Tha1, we evaluated the activity of the enzyme with other β-hydroxylated amino acids: L-threonine and L-allo-threonine (Fig. 5d). The enzyme showed activity on both L-threonine and L-allo-threonine, but not with the D-enantiomers. However, the preferred substrate of Tha1 is HTML with a catalytic efficiency in the order of 104âsâ1 Mâ1, followed by L-allo-threonine (102âsâ1 Mâ1) and L-threonine (101âsâ1 Mâ1) (Fig. 5e). These results suggest that Tha1 has a catalytic preference for β-hydroxylated L-amino acids with the erythro configuration. With respect to L-allo-threonine, the reaction with HTML has a similar kcat but a 50-fold lower Km (Supplementary Fig. 15j, c), suggesting a higher affinity for the intermediate of the carnitine pathway. The two human SHMTs have a similar a preference for substrates with the erythro (S,S) configuration, but are much more efficient with L-allo-threonine (~104 for SHMT1, ~102 for SHMT2) than with HTML (Fig. 5e; Supplementary Fig. 15a, b; Supplementary Table 4), which possesses a bulkier side chain (Fig. 5d).
To verify if the preference of Tha1 for the HTML substrate is a feature of threonine aldolase proteins of organisms with the carnitine biosynthesis pathway, we tested the activity of the low-specificity threonine aldolase eTA51 from E. coli, which, like other bacteria, does not have carnitine biosynthesis. Recombinant eTA was produced in intact form in the homologous host. Characterization of its catalytic efficiency for L-allo-threonine and HTML, showed high activity with both substrates with a slight preference for L-allo-threonine (Supplementary Fig. 15h, d).
HTML is a competitive inhibitor of KYAT1
Although KYAT1 is unable to catalyze the aldol cleavage on HTML, the good binding energies obtained with the screening suggest potential binding at the active site. We thus wanted to test if HTML can inhibit KYAT1 activity on L-kynurenine.
In the presence of an α-keto acid, L-kynurenine is converted by KYAT1 to the corresponding keto acid (4-(2-aminophenyl)â2,4-dioxobutanoate), which rapidly cyclizes to kynurenic acid (Supplementary Fig. 17a). By measuring the spectrophotometric signal at 310ânm of the final product, we were able to observe the progress of the reaction in the absence and in the presence of HTML (Supplementary Fig. 17b, c). After the addition of 0.5âmM of HTML to the reaction mixture, a slowdown of the reaction is observed (Supplementary Fig. 17c), suggesting an inhibitory action. We characterized the initial velocity of kynurenine transamination with increasing concentrations of HTML. The Lineweaver-Burk double reciprocal primary plot shows a family of straight lines intersecting on the y axis, typical of competitive inhibition with a constant Vmax and an increasing apparent Km (Fig. 5f). A Ki value of 4âmM was determined by the secondary plot (Supplementary Fig. 17d).
Crystal structure of mouse Tha1 improves HTML-OSMES results
Although the AlphaFold models in our screening are of high quality overall, there is a disparity in the dataset as evidenced by the different RMSD (root-mean-square deviations) with respect to the templates used for oligomer reconstruction (Supplementary Fig. 18). These differences depend on the availability of experimental structures from the same or closely related species. For instance, in the case of KYAT, SGPL, and SHMT, PDB structures are available from various mammals, including humans52,53,54,55 and mouse56,57, whereas in the case of Tha1, only PDB structures from distant bacterial homologs are available51,58. To verify if the results of our screening for Tha1 are confirmed or improved with the availability of an experimental structure, we decided to determine the crystal structure of mouse Tha1.
Mouse Tha1 crystallizes in two space groups, in orthorhombic F222 and in monoclinic C2, with one molecule and two molecules in the ASU, respectively. The PLP cofactor is visible only in the monoclinic structure; however, the active site is very similar in the two cases, with only minor differences. The expected tetrameric quaternary structure is formed by crystallographic symmetries, with four identical units in F222 (related by a 222 symmetry) and two identical dimers in C2 (related by a two-fold axis). The RMSD values between the single units (around 0.26â0.28âà , Supplementary Table 5) indicate that the monoclinic and orthorhombic structures are similar. Also the tetrameric assembly is conserved in the two space groups, with two main interfaces (Fig. 6a). As indicate by data from PISA analysis (Supplementary Table 6), the interface between units A and B (analogous to that between units C and D, termed âmain interfaceâ) is contributing stronger to the stability of the quaternary structure in comparison with the interface between units A and C (analogous to that between units B and D, termed âsecondary interfaceâ). Hence, the tetramer can be considered a dimer (ABâ+âCD) of dimers (Aâ+âB and Câ+âD), with the first dissociation being ABCD to ABâ+âCD (as determined by PISA). A comparison with the structure of the Thermotoga maritima threonine aldolase (PDB code 1M6S) returned RMSD values between 1.03 and 1.17âà for the single units (Supplementary Table 5), indicating a significant structure difference even though the secondary structures and the whole quaternary assembly are conserved. The major structural difference is related to an insertion of 10 residues in Tha1 between positions 337â346. In the two enzymes the position of the PLP cofactor is essentially conserved (Fig. 6b). In Tha1, the PLP cofactor, bound to Lys242, is stabilized in the active site by a network of hydrogen bonds and salt bridges with the side chains of Asp211, Arg214 and Thr98 from the same unit, and of Lys267 and Arg274 from the adjacent unit (Fig. 6b). His123 is making an aromatic stacking interaction with the PLP pyridine ring with a relative distance between the rings of 3.7âà . While the main interface has similar characteristics for the mouse and the T. maritima enzymes (as deduced by PISA analysis, see Supplementary Table 6), the secondary interface shows a higher degree of variability. Despite with similar buried area values, around 990â1060âà 2, it stronger contributes to the stability of the tetrameric assembly in the T. maritima enzyme, with much higher values in ÎGint (the solvation free energy gain upon formation of the assembly), ÎGint P-value, and the Complexation Significance Score (CSS). The secondary interface has a more hydrophobic nature in the T. maritima enzyme while it is more polar in the mouse enzyme. As a consequence, the tetrameric assembly of the mouse enzyme has a lower stability, with a ÎGdiss (the free energy of assembly dissociation) value of 5â6âkcal/mol compared to the 40.9âkcal/mol of the bacterial enzyme (for the ABCD to ABâ+âCD dissociation). We performed SEC-SAXS experiments that show the presence of a single component with a MW compatible to that of the sum of 4 units, indicating that the mouse enzyme, despite the lower stability, is tetrameric in solution (Supplementary Fig. 19).
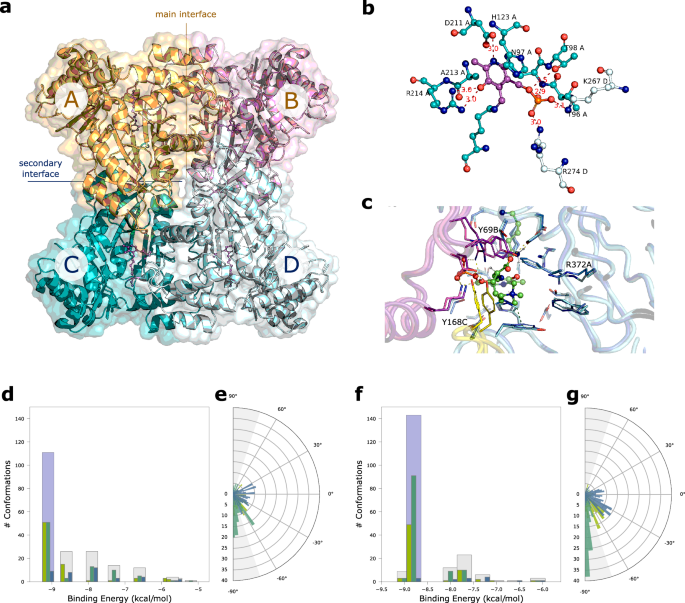
a Quaternary assembly of Tha1. The four PLP cofactors, one for each unit, are shown in violet ball-and-stick. The main interface, between units A and B (orange/magenta) and the secondary interface between units C and D (teal/pale cyan) are indicated. b Active site of Tha1. The main polar interactions of the PLP cofactor (violet) at the interface between subunits A (carbon atoms in teal) and B (carbon atoms in pale cyan) are indicated. Distances in à . c Comparison of AlphaFold model (dark colors) and the Tha1 crystal structure (orthorhombic F222, light colors) docked with HTML-PLP substrate. Different chains are colored in different colors. Non-carbon atoms are colored according to CPK convention. HTML-PLP and flexible catalytic lysine are shown in ball-and-sticks. The binding site residues (â¤4.5âà from HTML-PLP) are shown in sticks. Polar interactions are indicated with orange dashes, cation-Ï interactions are indicated with olive dashes; residues showing a different position in the model and crystallographic structure are labeled. Clustering of the HTML-PLP conformations at the Tha1 active site obtained with HTML-OSMES applied to AlphaFold model (d, e) or the crystal structure (f, g). Bar plots show the distribution of Ï1 (blue), Ï2 (emerald) and Ï3 (kiwi) angles in each cluster, with the catalytic cluster highlighted in light blue. Circular plots show the cumulative distribution of the three Ï angles for all clusters. |sin(Ï)| â¥0.95 values are defined by gray areas. Source data are provided as a Source Data file.
We repeated the docking screening by including in the data set the crystallographic structure of Tha1. The HTML-OSMES results show an increase in CFCs compared with what was obtained with the AlphaFold model. In the catalytic cluster, there are 91 CC-CFC within it, compared with 51 in the previous analysis (Fig. 6d, f; Supplementary Table 7). By comparing the two structures, some differences are observed in the side chains of the substrate binding residues (Fig. 6c). There are minor differences in the chain containing the catalytic lysine (e.g. Arg372A), while differences in the position of the residues contributed by the other chains (Tyr168C, Tyr69B) are more pronounced, suggesting that they result mainly from subunit assembly. Most importantly, it is observed that many more conformations of the entire docking analysis with the crystal structure have the relevant bond nearly perpendicular to the plane of the PLP (0°) (Fig. 6e, g), most of which have |sin(Ï2)|ââ¥â0.95 (gray area). The number of CC-CFC obtained by HTML-OSMES with the experimental structure would have allowed Tha1 to place second in the mouse ranking. Although to a lesser extent, an increase of CC-CFC value was also obtained by the AlphaFold model59 built with the addition of the Tha1 experimental structure as template (Supplementary Fig. 20; Supplementary Table 7).
Extension of the OSMES procedure to other enzymes
To test the possibility of extending the OSMES procedure to a different group of enzymes, we decided to apply OSMES to aldehyde dehydrogenases (EC: 1.2.1.-), a numerous protein family sharing a common catalysis mechanism (Supplementary Fig. 21). A member of aldehyde dehydrogenases, TMABADH, is functionally related to HTMLA, as it catalyzes the subsequent reaction in the biosynthetic pathway (see Fig. 3a). Also in this case we modeled an initial step of the reaction mechanism involving the nucleophilic attack by an active site cysteine to the substrate aldehyde carbon (Supplementary Fig. 21a), with the formation of a covalent intermediate60. The subsequent transfer of a hydride ion (Hâ) from this thioester intermediate results in the reduction of NAD(P)+ to NAD(P)H.
We considered substrate orientations in the active site as CFC when the distance between the aldehyde carbon and the catalytic cysteine thiolate was â¤3.5âà , which is regarded as an upper limit for near attack conformation60,61. Using this condition, we applied OSMES to human and murine aldehyde dehydrogenases encompassing two different PFAM domains (Gp_dh_N: PF00044 and Aldedh: PF00171), with a total of 20 and 22 enzymes for human and mouse, respectively (Supplementary Table 10). Since in these proteins the active site is enclosed within monomeric units, the oligomerization step was not performed.
Six different aldehyde molecules, which are known substrates of eight different enzymes, were used as positive controls for validation (Supplementary Fig. 21b). We observed a performance similar to PLP-dependent enzymes, with CC-CFC as the best ranking method (AUROC scoreâ=â0.86) and the energy-based methods (LCE, BCE) as a close second best. Converversely, the ranking methods based on the number of conformations (LCC, BCC) seem to lack discriminative power (Supplementary Fig. 21c, d). In various instances the conformation of the best cluster (BC) corresponded to a catalytically favorable conformation (Supplementary Fig. 21e).