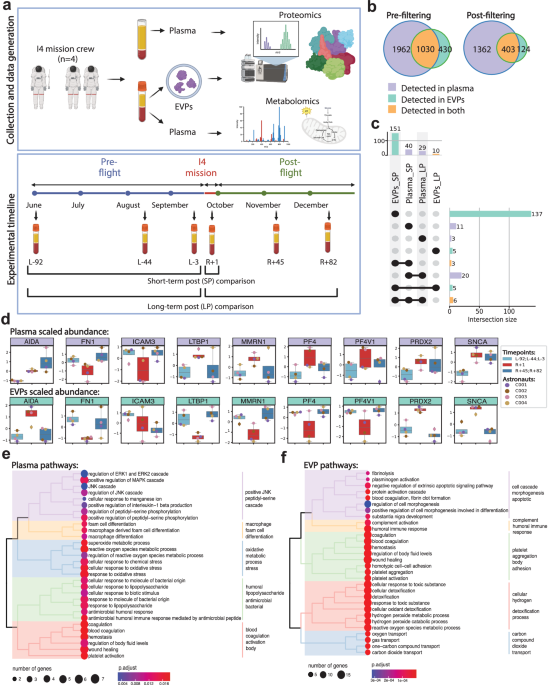
Changes in the proteomic profile of plasma and EVPs after 3-day spaceflight
To gain insight into secretome changes after 3 days of spaceflight, we profiled the plasma EVP proteins and plasma metabolites of the four i4 mission crew members (Fig. 1a) at three pre-launch dates (L-92, L-44, and L-3) and three post-flight timepoints upon return to Earth (R + 1, R + 45, R + 82) (Fig. 1a). For plasma proteomics, plasma was isolated using Cell Preparation Tubes (CPT) and processed with Seer’s 5-nanoparticle Proteograph assay23, while plasma EVPs were isolated as previously described24,25, and proteins were analyzed by nano-LC-MS/MS (Fig. 1a). Plasma metabolites were extracted using Aqueous Neutral Phase (ANP) hydrophilic and C18 hydrophobic liquid chromatography, and metabolites were identified and quantified by MS (Fig. 1a). We identified a total of 2,992 unique plasma proteins and 1,443 unique EVP proteins, with an overlap of 1,030 proteins shared by plasma and EVPs (Fig. 1b). These shared proteins are likely plasma EVP proteins as well as proteins that can be either soluble/free or EVP-associated. Plasma and EVP proteins were then filtered based on the number of not-detected (NAs) so that at least one condition has no missing data and coefficient of variation (<0.5), with 1,765 plasma circulating proteins remaining in plasma and 527 in EVPs (Fig. 1b).
a Overview of study design, sample collection, and processing of plasma and EVP proteomics. b Venn diagram of proteins measured in plasma and EVP, before (left) and after (right) filtering, based on the coefficient of variance, low abundance, and number of not assessed (NAs). c Upset plot showing the overlap of differentially abundant proteins (adjusted p-value < 0.05, |logFC | >1) across the different comparisons performed in plasma and EVPs. Differential abundance analysis was performed with limma with the following model ~astronaut+flightSatus and p-values have been adjusted to control the false discovery rate d Boxplots of the scaled abundance of the 9 proteins differentially abundant in both plasma and EVPs. Where available, data represents n = 4 astronauts averaged at the indicated condition (preflight and long-term postflight). Plasma data is the average of two technical replicates, EVP data represents one technical replicate per astronaut and timepoint. Boxes show the quartiles of the dataset while the whiskers extend to show the rest of the distribution except for outliers. e Gene Ontology enrichment was performed using clusterProfiler::enrichGO() on differentially abundant proteins in plasma (adjusted p-value < 0.05, |logFC | >1) at R + 1 vs. Preflight. Biological processes (BP) were selected, and treeplot was used to organize significant pathways (adjusted p-value < 0.05) into biologically relevant clusters. f Gene Ontology enrichment was performed using clusterProfiler::enrichGO() on differentially abundant proteins in the EVPs (adjusted p-value < 0.05, |logFC | >1) at R + 1 vs. Preflight. Biological processes were selected, and treeplot was used to organize and cluster the significant pathways (adjusted p-value < 0.05) into biologically relevant clusters. Source data are provided as a Source Data file.
To profile acute and long-term changes in the secretome after 3-days of spaceflight, we performed two comparisons: 1) immediately post-flight (R + 1) vs. all pre-flight (L-92, L-44, L-3) timepoints, as a measure of acute changes (short-term postflight, or SP) and 2) all post-flight (R + 1, R + 45, R + 82) vs. all pre-flight (L-92, L-44, L-3) timepoints, representing long-term changes (long-term postflight, or LP). Interestingly, even though fewer unique proteins were detected in EVPs, we identified more differentially abundant proteins (DAPs) in EVPs compared to plasma (151 DAPs in EVPs vs. 40 DAPs in plasma) at R + 1 (Fig. 1c, Supplementary Fig. 1). Importantly, the majority of EVP DAPs returned to pre-flight levels over time, with only 10 EVP DAPs (6.62%) remaining differentially abundant long-term post-flight (Fig. 1c). However, most plasma DAPs (72.5%) remained differentially abundant at the last timepoint (R + 82) (Fig. 1c), indicating a greater degree of recovery, and longer duration, than the EVP DAPs.
In addition, the EVP and plasma proteomes provided distinct information about spaceflight-associated changes. Specifically, 9 DAPs were shared between EVPs (5.96%) and plasma (22.5%) immediately post-flight (Fig. 1c). The 9 overlapping DAPs were Platelet factor 4 (PF4), Latent-transforming growth factor beta-binding protein 1 (LTBP1), Platelet factor 4 variant 1 (PF4V1), Alpha-synuclein (SNCA), Peroxiredoxin 2 (PRDX2), Fibronectin 1 (FN1), Axin interactor, dorsalization associated (AIDA), Multimerin 1 (MMRN1), and Intercellular adhesion molecule 3 (ICAM3) (Fig. 1d). Of these, PRDX2 and SNCA increased, while ICAM3 decreased in both EVPs and plasma. Since PRDX2 is an antioxidant enzyme26, its elevation in EVPs and plasma at R + 1 may indicate elevated oxidative stress. Increased SNCA level in the blood, including in EVPs, is a potential indicator of brain inflammation and stress27,28. Also of note, ICAM3 downregulation may reflect impairments in T cell-Dendritic cell (T-DC) interactions and immune function, as ICAM3 is crucial for the initial interaction between these two immune cells29. In addition, proteins associated with wound healing and coagulation, including PF4, PF4V1, and LTBP1, were increased in the plasma, but decreased in EVPs, potentially as a consequence of EVP capture in clots associated with spaceflight-induced thrombosis (Fig. 1d and Supplrmentary Fig. 1)14,15. However, PF4 and PF4V1 levels quickly returned to baseline, suggesting that the pro-thrombotic effect of spaceflight is temporary and reversible; these shifts of PF4 in plasma and EVPs were confirmed by ELISA (Supplementary Fig. 2a, b).
To gain a functional understanding of plasma and EVP proteome changes after spaceflight, we performed biological pathway enrichment analysis for the R + 1 DAPs in both plasma and EVPs. Interestingly, though individual DAPs mostly differed in plasma relative to EVPs, pathways enriched in these DAPs showed a consistent profile in plasma and EVPs. DAPs involved in reactive oxygen species (ROS) production, oxidative stress, wound healing, coagulation, immune function, and hemostasis pathways were enriched in both plasma and EVP profiles (Fig. 1e, f). These findings indicate that the plasma secretome reflects the hematologic changes (hemostasis, wound healing, coagulation), immune response/inflammation changes, and molecular changes in ROS metabolism after the 3-day spaceflight. We also note the increased abundance of several proteins related to the complement pathway, such as FCN3 which remains upregulated even at R + 45. This was validated in EVPs by western blotting (Supplementary Fig. 2c).
Changes in the metabolic profile of plasma after 3-day spaceflight
To capture spaceflight-related metabolic changes, we next profiled 1,135 metabolites in the plasma of the i4 crew using ANP hydrophilic and C18 hydrophobic liquid chromatography coupled with mass spectrometry30. Differential analyses of the metabolomics data identified a variety of metabolic pathways affected by spaceflight, with 100 differentially abundant metabolites (DAMs) identified when comparing the pre-flight (L-92, L-44, L-3) to immediately post-flight (R + 1) timepoint (Fig. 2a, b). Notably, none of these DAMs remained differentially abundant at timepoints after R + 1, indicating that these metabolic changes are acute, and also that metabolic homeostasis is restored rapidly upon return to Earth.

a Pie charts showing pathway annotations of differentially abundant metabolites (adjusted p-value < 0.05, |logFC | >1) at R + 1 vs. Preflight. Gray portions of the pie chart represent measured but insignificant metabolites in the specified category, while colored portions are labeled by metabolite pathway name and frequency. b Volcano plot of metabolites based on differential abundance (logFC>1 in red, logFC < −1 in blue) at R + 1 vs. Preflight. Labeled points are differentially abundant metabolites with connections to spaceflight-related anemia, inflammation, and oxidative stress. c Box plots of the scaled abundance of select differentially abundant metabolites associated with purine metabolism, glycerophospholipid metabolism, and anemia and hemolysis. Data represents n = 4 astronauts averaged at the indicated condition with one technical replicate per astronaut at each timepoint. Boxes show the quartiles of the dataset while the whiskers extend to show the rest of the distribution except for outliers. d Diagram of the sphingomyelin cycle, which is enriched in spaceflight-affected metabolites. Annotation boxes represent metabolites significantly changed at R + 1 vs. Preflight, with increased abundance indicated in red, decreased abundance in blue, and no significant change in gray (only boxed metabolites were measured). The dashed boxes indicate significant changes in some, but not all, chain lengths of the indicated lipid species. e Violin plots showing the results of Thiobarbituric acid reactive substances (TBARS) assay performed on astronaut plasma. Repeated measures one-way ANOVA was performed to assess significance. Source data are provided as a Source Data file.
Of the metabolites affected immediately post-flight, those involved in the purine metabolism pathway showed systematically increased abundance. Inosine, the metabolite with the largest positive fold-change (Fig. 2b), together with its precursor metabolite, purine, and its post-degradation metabolites, xanthine, and hypoxanthine, are all components of purine metabolism (Fig. 2c). We found that many of the most significantly altered metabolites belonged to the glycerophospholipid metabolism pathway (Fig. 2a), likely driven by a decreased abundance of lysophospholipid (LysoPC) and phosphatidylcholine (PC) species such as methylcarbamoyl platelet-activating factor (PAF) C-16 and LysoPAF C-16 (Fig. 2b, c)31. Accompanying these changes, many metabolites involved in the sphingomyelin cycle were differentially abundant, including an increase in uridine diphosphate (UDP), some sphingomyelin and glucosylceramides (GlcCer) species, and a decrease in sphingosine-1-phosphate (S1P) (Fig. 2c, d).
Since phosphatidylcholines (PCs) are the most abundant phospholipid species in cellular membranes, and membrane stability is disrupted by lipid peroxidation, we hypothesized that the decrease in PCs and LysoPCs (Fig. 2d) was indicative of lipid peroxidation secondary to spaceflight-induced production of free radicals32,33. The lipid peroxidation cascade generates a number of different intermediates depending on the target lipid species34. Malondialdehyde (MDA) is one of the major byproducts of lipid peroxidation and widely used as a biomarker of oxidative stress34. Therefore, to measure the overall lipid peroxidation in plasma, we quantified the levels of MDA adducts with thiobarbituric acid (TBA) using the thiobarbituric acid reactive substance assay (TBARS assay) (Fig. 2e), which revealed that lipid peroxidation was significantly increased (p-value = 0.0013) immediately post-flight (R + 1) and returned to baseline levels after several weeks (R + 45).
Upregulation in production of antioxidants in the plasma and EVPs in response to spaceflight
We next investigated the common signature of oxidative stress and cellular detoxification found in EVP and plasma DAPs35,36 (Supplementary Fig. 3). Specifically, ROS scavenging is dependent on ROOH and H2O2 detoxification via superoxide dismutase (SOD1, SOD2), catalase (CAT), and peroxiredoxins37,38. Compared to ground controls, plasma from post-flight astronauts displayed an upregulation of antioxidant proteins and a distinct metabolic profile retained at both the immediate and long post-flight timepoints (Fig. 3a). Antioxidant proteins were also significantly enriched in EVPs in the intermediate post-flight timepoint, along with metabolic proteins involved in anabolic metabolism and cell growth, which may improve donor cells’ antioxidant capacity and cell bioenergetics36,39,40,41. Our findings indicate the body upregulates the production of anabolic metabolism and antioxidants in the plasma and EVPs, especially in the immediate-post-flight timepoint, likely to compensate for increased ROS (Fig. 3a). At the longterm post-flight timepoint, these protein levels are abolished in the EVPs, but not in the plasma, which maintains an upregulation of antioxidant proteins and an altered metabolic profile. These data indicate that intracellular ROS levels and metabolic profiles can remain differentially abundant following spaceflight for at least 80 days after landing.

a The top (plasma) indicates significantly differentially abundant proteins in the plasma and EVPs from immediate or long-term post-flight or both immediate and long-term post-flight groups compared to ground controls. Antioxidant proteins are white-colored, and proteins involved in mitochondrial metabolism are orange. The purple edges represent upregulated proteins, and the green edges represent downregulated proteins. The lower panel (inside the cell) shows the antioxidant and mitochondrial metabolism protein functions within the cell. b Overview of the bipartite correlation network with proteins and metabolites as nodes. Ellipses depict proteins, and metabolites are represented by square nodes. The edges indicate significant correlations between the nodes. A solid line indicates a positive correlation, while a dotted line indicates a negative correlation. Nodes are colored based on log2-fold changes immediately post-flight compared to pre-flight time points. c The antioxidant defense subnetwork is enriched in peroxidases, antioxidant enzymes, and antioxidant molecules, indicating activation of extensive antioxidant response. d The immunosuppression and anti-inflammatory response subnetwork is enriched in anti-inflammatory molecules and protein markers of immune cells that lower post-flight, indicating a deregulated immune response. Source data are provided as a Source Data file.
Integrating proteomics and metabolomics reveals a common signature of antioxidant defense and immune dysregulation
To investigate the molecular processes at the interface of altered proteins and metabolites, we undertook a correlation-based, integrated approach. Specifically, we correlated the proteins and metabolites across all timepoints, using the annotated DAPs and DAMs altered immediately post-flight (R + 1) compared to pre-flight (L-92, L-44, L-3). As a result, we identified 26 significant (False Discovery Ratio (FDR) < 5%) correlations between plasma proteins and plasma metabolites. In contrast, 1,416 correlations were significant between EVP proteins and plasma metabolites (Fig. 3b). The significantly correlated molecules were visualized as a network with proteins and metabolites as nodes, and the correlation between them as edges. Among these changes, two of the most commonly observed molecular changes post-flight were oxidative stress and immune dysregulation, as discussed below1,22,42,43,44,45,46,47.
First, we analyzed the interface between the ROS pathway and metabolites by correlating the proteins in the ROS pathway with metabolites. Exposure to radiation, microgravity, and hypoxia during spaceflight all induce the production of free radicals leading to oxidative stress, which can impact on cardiovascular, immune, neurological, and metabolic systems1. The ROS subnetwork from the i4 crew consisted of 26 nodes, including 6 proteins and 20 metabolites, with 38 correlation-based edges between the metabolites and proteins (Fig. 3c). All 6 proteins (100%) and 12 metabolites (60%) were lower post-flight, and 8 metabolites (40%) were higher post-flight. Within this subnetwork, three antioxidant enzymes from the peroxiredoxin family (PRDX1, PRDX2, PRDX6) which scavenge peroxides within cells48, were increased immediately post-flight. In addition, three other enzymes that degrade ROS, namely SOD1, CAT and glutamate-cysteine ligase (GCLC), were also increased post-flight. SOD1 catalyzes the conversion of superoxide into hydrogen peroxide, which CAT can then degrade49. GCLC is a rate-limiting enzyme for the de novo synthesis of glutathione, a widely studied antioxidant that maintains the cellular redox balance50. Moreover, antioxidants, including inosine and taurine, were significantly increased immediately post-flight (q-value < 0.05). In addition to its antioxidant capacities, inosine dampens cytokine production, normally ameliorating inflammation51,52,6,53. Taurine is also an antioxidant that protects immune cells during oxidative stress54, and its upregulation suggests that immediate post-flight antioxidant production compensates for spaceflight-induced oxidative stress.
We next analyzed the interface between the immune system and metabolism by correlating immune cell markers with specific metabolites. The immune subnetwork consisted of 56 nodes, including 10 proteins and 46 metabolites, with 95 correlation-based edges between the metabolites and proteins (Fig. 3d). All 10 proteins (100%) and 33 metabolites (71.7%) were lower immediately post-flight, and 13 metabolites (28.3%) were higher immediately post-flight. Within this subnetwork, all the protein markers of immune cells were decreased immediately post-flight. Moreover, our analysis indicated that anti-inflammatory and antioxidant molecules, namely inosine, purines, and taurine, were increased immediately post-flight. While taurine and inosine are antioxidants51,54, purines (e.g., adenosine) modulate the immune system by inhibiting the production of pro-inflammatory cytokines and free radicals51. In addition, 4-aminobutanoate (GABA), an immune-modulatory neurotransmitter, was also increased immediately post-flight, which can inhibit cytokine production55,56. This subnetwork may reflect widespread inflammation preceding the post-flight immunosuppression and anti-inflammatory responses, consistent with some of the NASA Twins Study results22 and studies of physiological stress, radiation, altered circadian rhythm42,43, and reactivation of latent herpes viruses44,57.
Immune cells contribute to the observed secretome changes after spaceflight
To delineate the contribution of immune cells to the secretome, we compared our proteomic data to single nuclei gene expression of peripheral blood mononuclear cells (PBMCs) from the i4 crew using the 10X Genomics single-cell multi-ome kits for epigenetic and gene expression profiling (see Methods). Of the ~30,000 genes detected in PBMCs, 273 genes were detected in both plasma and EVPs (Fig. 4a). In addition, 1131 genes were uniquely reflected in the plasma proteome while 163 genes were uniquely found in the EVP proteome (Fig. 4a).
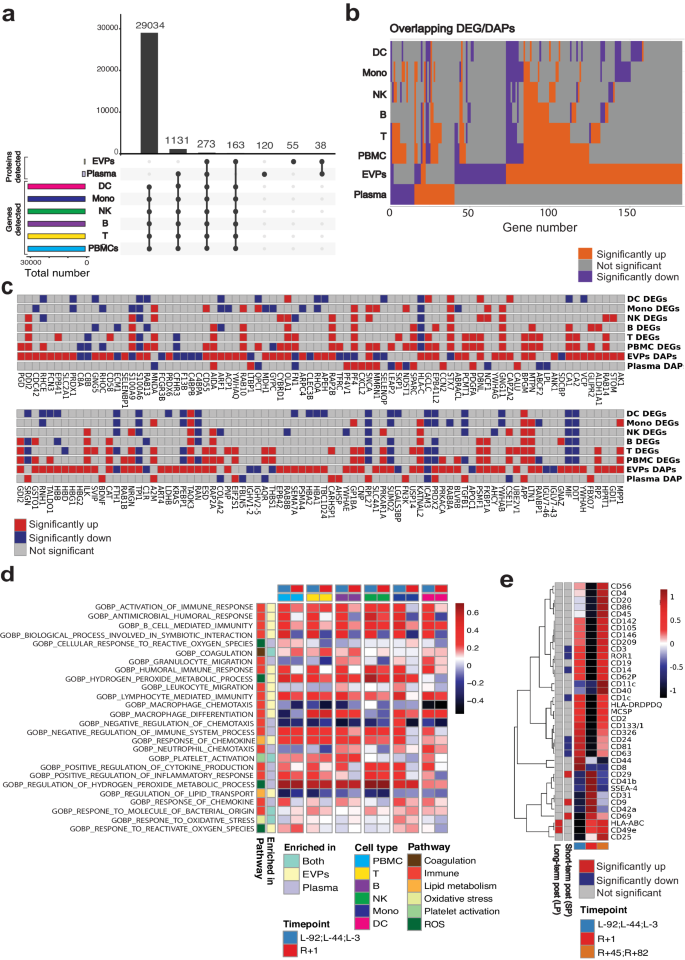
a Upset plot of identified genes in PBMC and identified proteins in EVPs and plasma. b Overlap of immune cell DEGs, plasma DAPs, and EVP daps. Up-regulated genes/proteins (Immune cells: p-value < 0.05, secretome: adjusted p-value < 0.05, fold change > 0) are depicted in orange. Down-regulated genes/proteins (Immune cells: p-value < 0.05, secretome: adjusted p-value < 0.05, fold change <0) are depicted in colored purple. Non-significant genes/proteins are depicted in colored gray. Wilcoxon rank sum tests were performed. c Expression of secretome DAPs in immune cells. d Fold change (R + 1/pre-flight) of the selected secretome-enriched pathways normalized score in immune cells. Among the secretome-enriched pathways, immune function, oxidative stress, antioxidant, lipid metabolism, coagulation, and platelet activation pathways were selected. e MACSPlex analysis of immune marker expression in EVPs. Source data are provided as a Source Data file.
When comparing the differentially expressed genes (DEGs) of PBMCs with the plasma DAPs and EVP DAPs (Fig. 4a), we found that 12 (30%) plasma DAPs and 27 (17.8%) EVP DAPs were also differentially expressed in the PBMCs. Of those overlapping DAPs, 6/12 were differentially abundant in the same direction in both plasma and PBMCs. Additionally, 14/27 of the overlapping EVP DAPs were differentially abundant in the same direction in both EVPs and PBMCs. This likely indicates that circulating and EVP proteins reflect gene expression changes in immune cells, while validating that secreted proteins can also originate from non-immune cells and distal organs.
To further examine the connection between the EVP proteome and the PBMCs transcriptional states, we examined the overlap between EVP DAPs and PBMC DEGs. The crew EVP profiles showed a higher overlap with the DEGs among lymphoid cells (T cell, B cell, Natural Killer (NK) cell) than with myeloid cells (Fig. 4b), and the same trend was observed for plasma DAPs (Fig. 4b), indicating that lymphoid cells contributed more than myeloid cells to the observed changes in the secretome. Among the genes differentially abundant in EVPs and PBMCs, we noted several immune genes such as Integrin linked kinase (ILK), which was upregulated in EVPs and PBMCs, while carbonic anhydrase 8 (C8A) and complement C8 beta chain (C8B) were downregulated in EVPs and PBMCs (Fig. 4c).
Among the antioxidant and oxidative stress-related proteins, PRDX2 was again upregulated in both plasma and EVPs, but was downregulated in DC cells, Monocytes, T cells and B cells, indicating that the upregulation of PRDX2 and EVPs seen in plasma does not originate from immune cells (Fig. 4c). To disentangle the relationship between immune cells and the pathways enriched based on the secretome DAPs, we calculated the fold changes of normalized enrichment score (NES) of immune cells at R + 1 versus pre-flight for the significantly enriched secretome DAPs pathways related to coagulation, immune function, lipid metabolism, oxidative stress, platelet activation, and reactive oxygen stress (Fig. 4d and Supplementary Fig. 3). We found an enrichment in oxidative stress and ROS pathways in T cells, NK cells, monocytes, and DC at R + 1 compared to pre-flight timepoints.
To gain insight into potential cellular sources of plasma EVPs, we used the MACSPlex Exosome profiler, which estimates the abundance of EVs expressing one of 37 markers specific for various immune cell types. We found significant increases in the pan-EVP marker CD958,59, integrin beta-1 (ITGB1, also CD29)60, and B/T activation marker (CD69)61 and significant decreases in Alveolar Type I/Brain (Receptor tyrosine kinase like orphan receptor, ROR1)62,63,64, melanocytes (Melanoma chondroitin sulfate proteoglycan, MCSP)65,66, T cells (CD3)67, additional pan-EVP markers (CD6368, CD81), B cell (CD24)69, and DC (CD1C)70 markers (Fig. 4e). The increase in CD9 at R + 1 correlates with an overall increase in EVP production post-flight15, while the decrease in CD63 and CD81 at R + 1 is consistent with the increase in CD9+ EVPs produced by platelets involved in coagulation. Of note, CD69 (a T and B cell activation marker) was increased at R + 1, consistent with inflammation revealed by the other omics analyses. The increase in ITGB1 (CD29), which complexes with integrin subunit alpha 5 (ITGA5, also CD49e)71, a heterodimer expressed on activated lymphocytes, endothelial cells (ECs), osteoblasts and which binds fibronectin and L1 cell adhesion molecule (L1CAM, a central nervous system axonal protein), may be consistent with vascular permeability, and systemic inflammation. Several DC markers, CD24, CD1c, and CD209, were also decreased at R + 1, consistent with suppressed DC function.
We hypothesized that the significantly changed immune markers in EVPs would overlap with the i4 immune cell DEGs (Supplementary Fig. 4). Indeed, we found that while DEGs in PBMCs were driven more by T and B cells, all cell types showed changes in vesicle regulation (Supplementary Fig. 5). CD3 delta, a pan-T cell marker, was downregulated in PBMCs, T cells, and their EVPs, whereas a different T cell marker (CD69) was downregulated in cells, but enriched in EVPs. This could indicate that CD69 is selectively shuttled into EVPs, or selectively enriched in EVPs derived from tissue-resident memory T cells. Moreover, CD63 was consistently downregulated in innate immune cells (DC, NK, macrophages) and EVPs immediately post-flight, indicating that cellular reduction was responsible for decreased EVP CD63 levels.
Red blood cells do not contribute to secretome changes after spaceflight
To delineate the contribution of blood cells to the secretome, we compared the gene expression profiles of whole blood direct RNA-seq data (Oxford Nanopore) from the i4 crew and identified 61 overlapping DEGs. We then compared the gene list with the plasma and EVP DAPs (Supplementary Fig. 6a, b). Interestingly, protein abundances in EVPs and gene expression in whole blood were inversely correlated (increased in EVPs and decreased in whole blood). The expression of these genes in PBMCs, however, was mostly aligned with the protein abundance, with at least one cell type showing a significant increase in PBMCs for SNCA. Of note, SNCA overlapped in whole blood DEGs, plasma DAPs, and EVP DAPs.
Overall, 8 genes were shared between whole blood DEGs and EVP DAPs and one gene was shared between blood DEGs and plasma DAPs. To determine the contribution of whole blood and PBMC to the secretome, we then examined expression of these overlapping DAPs (AHSP, AK1, ANK1, BLVRB, EPB42, HBD, ENBP1, SNCA) before and after spaceflight (Supplementary Fig. 6c), and analyzed this list for enriched gene functions. Significant enrichment (q < 0.01) was observed in heme metabolism, anemia, hematologic disease, brain function (terminal button, axon part)-related pathways (Supplementary Fig. 6d). The gene SLC4A1 was present in nearly all overrepresented groups (excluding the cell cortex and axon part), indicating a brain-related phenotype that warranted further investigation.
Brain-related signatures increased in the secretome after spaceflight
Spaceflight exposes the human brain to several stressors, which have the potential to cause short-term and long-term neurological effects, including SANS, body fluid shift, neuroinflammation, and neurodegeneration12,72,73,74. In addition, an increasing body of evidence suggests that the spaceflight environment could induce blood-brain barrier (BBB) disruption72,75,76,77,78. For example, EVP proteomic profiles of NASA Twins study obtained three years post-return from a year-long flight revealed brain-associated proteins in the plasma of the astronaut twin, but not the ground control twin15.
Since EVPs are known to be released from distal organs such as the brain, and could be detected in plasma, we examined post-spaceflight EVPs for any enrichment of brain-specific or brain-associated proteins. Indeed, both EVPs and plasma DAPs were enriched for brain function and brain injury-related pathways (Fig. 5a), including neurodegeneration pathways, neuron death, and amyloid fibril formation. Moreover, Gene Set Enrichment Analysis (GSEA) analysis revealed that brain-associated proteins were increased in plasma at R + 1 (Fig. 5b), which matches orthogonal data from a JAXA cfRNA-seq study that also revealed an increase in brain-enriched proteins immediately post-flight (R + 3) (Fig. 5c) (study OSD-530 of 6 Japanese astronauts on the International Space Station, ISS). Of note, the spike in brain signatures for plasma, cfRNA, and exosome proteins was most pronounced in the days after landing back on Earth (R + 1 and R + 3) (Fig. 5d).

a Overrepresentation analysis of significantly enriched pathways (adjusted p-value < 0.05) related to brain function and injury of EVPs and plasma DAPs at R + 1 (adjusted p-value < 0.05, Left: EVP, Right: plasma). b Gene set enrichment analysis (GSEA) of EVP DAPs immediately post-flight and long-term post-flight based on the tissue-enriched database derived from the Human Protein Atlas database. GSEA was performed with fgsea::fgsea() using minSize=5 and maxSize = 500 as parameters. Significant results (adjusted p-value < 0.1) are shown. c Gene set enrichment analysis of cfRNA measured immediately post-flight JAXA CFE mission based on the tissue-enriched database derived from the Human Protein Atlas database. GSEA was performed with fgsea::fgsea() using minSize = 5 and maxSize = 500 as parameters. Significant results (adjusted p-value < 0.1) are shown. d Abundance of brain-enriched proteins in EVPs. Data is from n = 4 astronauts, representing one technical replicate per astronaut and timepoint averaged at the indicated condition (preflight, and long-term postflight). Boxes show the quartiles of the dataset while the whiskers extend to show the rest of the distribution except for “outliers”. Source data are provided as a Source Data file. e Abundance of brain-enriched proteins in EVPs isolated from naive, ground control mice.
To examine possible sources of spaceflight-associated brain signatures in plasma, two hypotheses were examined: (1) proteins are purposely packaged and shuttled from the brain into EVPs and released or (2) BBB integrity is disrupted, indicating “leakiness” of brain proteins. To address the first hypothesis, we examined EVP protein cargo data in 13 mouse tissues from a public EVP atlas25 (blood, thymus, lymph node (LN), brown adipose tissue (BAT), bone, brain, heart, kidney, liver, lung, spleen, white adipose tissue, and muscle). Of the 16 brain-annotated proteins, 3 were brain-exclusive (CNP, EP41, NRGN), 2 were highly enriched in the brain (Epb4L1 and PRKACA), and five others were highest in the liver. However, SNCA, semaphorin 7 A (SEMA7A), and small VCP-interacting protein (SVIP) were not detected in mouse brain EVPs, nor in other tissues (Fig. 5e), indicating that packaging of these proteins into EVPs is unlikely, given their absence in the murine brain tissue.
To test the alternative hypothesis of BBB disruption, we examined the expression of biomarkers previously associated with BBB integrity, specifically S100 calcium binding protein B (S100B), Enolase 2 (ENO2) and Platelet Endothelial Cell Adhesion Molecule (PECAM-1)79,80 (Fig. 6a, b). While the proteins ENO2 and S100B showed no significant difference for pre/post-flight, PECAM-1 showed an increase in the plasma protein abundance at R + 1 in C001, C003, and C004, and a postflight increase as well when measured at R + 45 and R + 82 (Wilcoxson rank sum, p = 0.07)(Fig. 6a). To further examine the in vivo changes in PECAM-1, we used brain tissue from rodents flown on the RR-18 mission, which spent 35 days on the ISS. After spaceflight, the RR-18 rodents were returned to Earth, wherein the flight samples (FLT) were dissected and fixed onto slides for straining at the same time as the ground controls (GC). Interestingly, significantly increased PECAM-1 immunoreactivity (n = 5 replicates, two-sided Student’s paired t-Test, p = 0.023) was detected in the FLT samples relative to the GC group (Fig. 6b), implicating PECAM-1 as a possible spaceflight-related marker for BBB integrity.

a Abundance of blood-brain barrier (BBB) integrity peptides in plasma of i4 astronauts shown as violin plots. Data is from n = 4 astronauts. Each blood proteomic measurement was performed in two technical replicates per astronaut and timepoint. Displayed data represents the average of the technical replicates which were further averaged at the indicated condition (preflight, and long-term postflight). Boxes show the quartiles of the dataset while the whiskers extend to show the rest of the distribution except for “outliers”. b Representative images of hippocampal PECAM-1 in the flight (FLT) and ground control (GC) mice (n = 5). PECAM-1 positive cells were identified based on red fluorescence, while endothelium was stained with lectin (green). The nuclei were counterstained with DAPI (blue). In the control hippocampal region, few positive cells were found. In the hippocampal region of FLT mice, enhanced PECAM expression could be detected. There was a significant difference between FLT and GC groups with p < 0.05 (n = 5 replicates, two-sided Student’s paired t-Test, p = 0.023).Source data are provided as a Source Data file. Scale bar = 50 mm.