Donor samples
Anonymized whole blood samples were collected under informed consent from healthy volunteers through the Interregionale Blutspende (IRB) of the Swiss Red Cross (SRK) in Bern, Switzerland. Sample collection was approved by Ethikkommission Nordwest- und Zentralschweiz (EKNZ), approval number Req-2017-00050. Sex, gender, race, and population characteristics are not defined as the samples were anonymized and the research does not fall outside of Swiss Human Research Act. No ethics oversight is needed according to Swiss Law for this study.
Cell culture
THP1 cells were cultured using RPMI 1640 Medium, GlutaMAX⢠Supplement medium (Thermo Fisher Scientific, Cat#61870010) supplemented with 10% heat inactivated-FCS (Bioconcept, Cat#2-01F36-I), 25âmM Hepes (Thermo Fisher Scientific, Cat#15630080), 1âmM sodium pyruvate (Thermo Fisher Scientific, Cat#11360070), and 50âU/ml penicillin-streptomycin (Thermo Fisher Scientific, Cat#15140122). Cells were passaged every 4-â5 days by dilution to 0.2â0.3 million cells/ml.
Dual-THP1TM cells (Invivogen, Cat#thpd-nfis) were cultured using RPMI 1640 Medium, GlutaMAX⢠Supplement medium (Thermo Fisher Scientific, Cat#61870010) supplemented with 10% heat inactivated-FCS (Bioconcept, Cat#2-01F36-I), 2âmM l-glutamine (Thermo Fisher Scientific, Cat#25030081), 25âmM Hepes (Thermo Fisher Scientific, Cat#15630080), 1âmM sodium pyruvate (Thermo Fisher Scientific, Cat#11360070), and 50âU/ml penicillin-streptomycin (Thermo Fisher Scientific, Cat#15140122). Cells were passaged every 4â5 days by dilution to 0.2â0.3 million cells/ml.
HEK293T or HEK293-JumpIN cells were cultured with DMEM, high glucose, GlutaMAX⢠Supplement, pyruvate (Thermo Fisher Scientific, Cat#31966021) supplemented with 10% heat inactivated-FCS (Bioconcept, Cat#2-01F36-I), 10âmM Hepes (Thermo Fisher Scientific, Cat#15630080), and 50âU/ml penicillin-streptomycin (Thermo Fisher Scientific, Cat#15140122). Cells were passaged every 3â4 days, by diluting the cells 1 to 5, after releasing the attached cells using TrypLE (Thermo Fisher Scientific, Cat#12605010).
For transfection studies, the TransIT-LT1 transfection reagent (Mirus, Cat#MIR2304) was used according to the manufacturerâs protocol. For the six-well format (TPP, Cat#Z707759), 1 million HEK293T cells were seeded one day before transfection. For each well, 2.5âµg of DNA in 7.5âµl of TransIT-LT1 transfection reagent diluted in 250âµl optiMEM (Thermo Fisher Scientific, Cat#31985062). After 15âmin of incubation of the transfection reagent with DNA, the transfection mix was added dropwise to the cells. Then 48âh after transfection, cells were treated with the compound of interest.
For lentiviral particle production, 1âÃâ107 HEK293T cells were seeded on collagen-coated T75 flasks (Corning, Cat#356485) and then 24âh later, the cells were transfected with 1.84âµg of DNA of interest with 2.24âµg of ready-to-use lentiviral packaging plasmid mix (Cellecta, Cat#CPCP-K2A) using TransIT-LT1 transfection reagent (Mirus, Cat#MIR2304) according to the manufacturerâs protocol. Subsequently, the media was changed 24âh after transfection and lentivirus-containing supernatants were collected after 72âh. Finally, lentiviral particles were filtered using a 0.45âµm filter (Sartorius, Cat#16537), before concentrating 10-fold using the Lenti-X concentrator (Takara, Cat#631232) according to the manufacturerâs protocol.
Cell viability assay
Compound toxicity was measured by luminescence-based cell viability assay, CellTiter-Glo® Assay (Promega, Cat#G7570) was used according to the manufacturerâs instructions. Viability assays were performed in clear bottom 96-well plates (TPP, Cat#ZZ707902) where THP1-Cas9 cell cells were seeded at a cell density of 0.4 million cells/well and with the indicated doses of compounds. Cells were treated right after seeding. Luciferase measurement was performed after an indicated period (0â72âh) of compound incubation. To detect luciferase activity, CellTiter-Glo® Assay (Promega, Cat#G7570) was used according to the manufacturerâs protocol with minor changes. The reagent (50âµl) was added on top of the 50âµl cell-compound containing wells in a black with clear bottom 96-well plates (Greiner, Cat#655090) where the bottom of the plate was sealed (PerkinElmer, Cat#6005199). Luminescence was measured for 0.1âs with the EnVision® Multimode plate reader (Revvity, workstation version 1.14.3049.1193). Each of the experiments was repeated in three independent biological replicates. Fold increase in luminescence was calculated by dividing each read by its matching control group (DMSO treated), and results were plotted using GraphPad Prism 9.
Proteomics
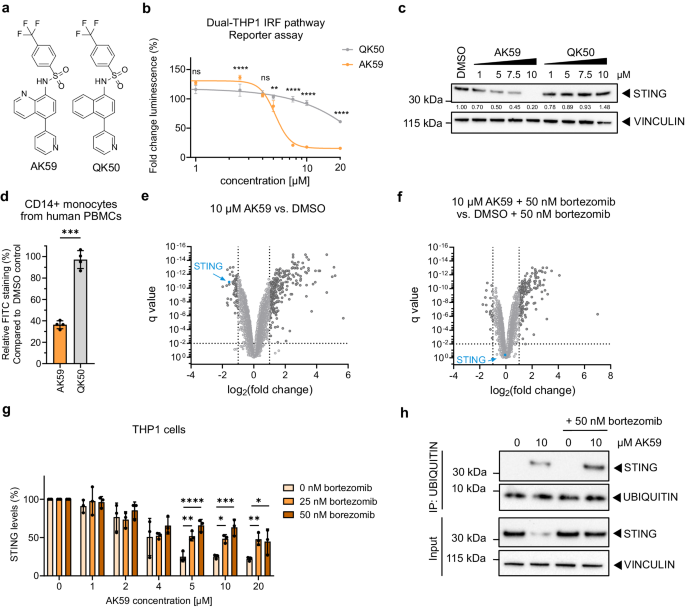
THP1-Cas9, Dual-THP1-Cas9, Dual-THP1-Cas9 STING KO cells or Dual-THP1-Cas9 HERC4 KO cells (2âÃâ106 per well in a six-well plate) were seeded and treated on the same day either with 0.1% (final) DMSO (Sigma-Aldrich, Cat#D8418) or 50ânM bortezomib (Sigma-Aldrich, Cat#504314, diluted in 0.1% DMSO final). Two to three biological replicates were used for proteomics. One hour after bortezomib treatments, 10âµM AK59 (in-house production, see Supplementary Methods) or 10âµM QK50 (in-house production, see Supplementary Methods) (0.1% (DMSO final) was added on top of the cells and incubated for 16âh. DMSO treatment was used as a control. Cells were collected and washed with 1x DPBS (Thermo Fisher Scientific, Cat#14190094) after compound incubation. Before proteomics analysis of the samples, the conditions were checked using western blot (for more details, see western blot) to confirm targeting effects on STING. PXD040291 dataset was run as 16 samples and PXD046677 run as 18 samples.
TMT-labeled peptides were generated with the iST-NHS kit (PreOmics, Cat#P.O.00030) and TMT16plex or TMT18plex reagent (Thermo Fisher Scientific, Cat#A44522, Cat#A52047) using around 2âÃâ106 cells per sample. Equal amounts of labeled peptides were pooled and ~300âµg of pooled peptides were separated on a Waters XBridge BEH C18, 130âà , 3.5âµm, 150âÃâ1âmm column with a gradient from 100% buffer A (10âmM ammonium formate in water, pH 11) to 55% buffer A and 45% buffer B (10% (v/v) 10âmM ammonium formate, pH 11 in water and 90% (v/v) acetonitrile) in 60âmin with a flow rate of 60âµl/min. Alternating rows of the resulting 72 fractions were pooled into 24 samples, dried, and resuspended in water containing 0.1% formic acid.
The LC-MS analysis was carried out on an EASY-nLC 1200 system coupled to an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). Around 1.5âµg of peptides from each sample (each condition in 2â3 biological replicates) were separated on a 25âcm long Aurora Series UHPLC column (Ion Opticks, Cat# AUR3-25075C18) with 75âµm inner diameter and a gradient from 95% buffer A (0.1% formic acid in water) and 5% buffer B (0.1% formic acid in 80% (v/v) acetonitrile and 20% (v/v) water) to 65% buffer A and 35% buffer B in 168âmin and then in 9âmin to 35% buffer A and 65% buffer B with a flow rate of 400ânl/min. MS1 spectra were acquired at 120k resolution in the Orbitrap, MS2 spectra were acquired after CID activation in the ion trap, and MS3 spectra were acquired after HCD activation with a synchronous precursor selection approach using 5 or 8 notches and 50âK resolution in the Orbitrap. LC-MS raw files were analyzed with Proteome Discoverer 2.4 (Thermo Fisher Scientific).
Briefly, spectra were searched with Sequest HT against the Homo sapiens UniProt protein database and common contaminants (2019, 21,494 entries). The database search criteria included 10 ppm precursor mass tolerance, 0.6âDa fragment mass tolerance, a maximum of three missed cleavage sites, dynamic modification of 15.995âDa for methionines, static modifications of 113.084âDa for cysteines, and 304.207âDa for peptide N-termini and lysines. The Percolator algorithm was applied to the Sequest HT results. The peptide false discovery rate was set to 1% and the protein false discovery rate was set to around 5%. TMT reporter ions of the MS3 spectra were integrated with a 20 ppm tolerance and the reporter ion intensities were used for quantification.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository51 with the dataset identifier PXD040291 (DOI 10.6019/PXD040291) and PXD046677 (DOI 10.6019/PXD046677). Protein relative quantification was performed using an in-house developed R (v.4.2) script, available on GitHub (https://github.com/Novartis/px_tmt_daa). This analysis included multiple steps; (1) data filtering (exclusion of peptides mapping to multiple proteins, exclusion of PSM where the number of SPS mass matches were <60%, the precursor interference was >50% or the average reporter ion s/n <10, as well as exclusion of PSMs with missing reporter ion signals); (2) global data normalization by equalizing the total reporter ion intensities across all channels, (3) summation of reporter ion intensities per protein and channel, calculation of protein abundance log2 fold changes (L2FC) and testing for differential abundance using moderated t-statistics52 where the resulting p values reflect the probability of detecting a given L2FC across sample conditions by chance alone. Subsequently, the p values were adjusted for multiple testing using the BenjaminiâHochberg method (q values).
Western blotting and co-immunoprecipitation
AK59 and QK50 compound treatments were always kept at 16âh incubation and 10âµM concentration unless otherwise indicated. Proteasomal inhibition (bortezomib Sigma-Aldrich, Cat#504314, diluted in 0.1% DMSO final, MG132 Selleckchem, Cat#S2619) or neddylation inhibition (MLN4924, Selleckchem, Cat#S7109) was initiated 1âh prior to compound treatment and maintained during the 16âh of AK59 incubation.
Cells were pelleted and washed with 1x DPBS (Thermo Fisher Scientific, Cat#14190094) before lysis. For western blot analysis, cell pellets were lysed with 5x extraction buffer (from co-immunoprecipitation kit, Thermo Fisher Scientific, Cat#14321D), diluted to 1x with 1x DPBS (Thermo Fisher Scientific, Cat#14190094) and supplemented with cOmplete protease inhibitor (Sigma-Aldrich, Cat#CO-RO) for 30âmin on ice with pulse-vortexing every 5âmin. To remove cell debris, lysate was spun at 15,000âÃâg at 4â°C for 15âmin. The supernatant containing proteins were collected in a fresh tube, and protein quantification was done using Pierce BCA assay (Thermo Fisher Scientific, Cat#23225) according to the manufacturerâs protocol. About 20âµg of protein/well in 20âµl loaded for each sample and then prepared with NuPAGETM LDS sample buffer (Thermo Fisher Scientific, Cat#NP0007) and NuPAGE sample reducing agent (Thermo Fisher Scientific, Cat#NP0004), boiled in 70â°C for 10âmin, loaded on pre-cast either 10-well, 12-well, or 15-well NuPAGETM 4 to 12% Bis-Tris 1.5âmm mini protein gel (Thermo Fisher Scientific, Cat#NP0335BOX, NP0322BOX, and NP0336BOX respectively). A protein ladder (Thermo Fisher Scientific, Cat#26619) was used to determine protein size. Samples run in 1x MES SDS running buffer (Thermo Fisher Scientific, Cat#NP0002) for 40â50âmin at 200âV. Semi-dry transfer was performed using Trans-blot turbo transfer (Biorad, Cat#1704156) with ready-made PVDF membrane-containing transfer packs according to the manufacturerâs protocol. Primary antibodies used in this study were all anti-human: STING (1:1000, CST, Cat#13647 or 1:500, Thermo Fisher Scientific, Cat#MA526030), UBIQUITIN (1:500, CST, Cat#3936), HERC4 (1:500, abcam, Cat#ab856732, batch number GR3184017-12), UBA6 (1:1000, CST, Cat#13386), UBA5 (1:500, abcam, Cat#ab177478), phospho-IRF3 (1:1000, abcam, Cat#76493), phospho-TBK1 (1:500, CST, Cat#5483), SMO (1:500, abcam, Cat#ab236465) and PGR (1:250, CST, Cat#8757). As loading control, ACTIN (1:500, Sigma, Cat#A5441), TUBULIN (1:500, CST, Cat#2146), and VINCULIN (1:500, CST, Cat#13901) were used, as described in figure legends. For secondary antibodies, HRP-conjugated anti-mouse (1:2500, CST, Cat#7076) and anti-rabbit (1:2500, CST, Cat#7074) antibodies were used. For detection, either Amersham ECL prime western blotting detection reagent (Cytiva Life Sciences, Cat#RPN2232) or SuperSignalTM West Femto reagent (Thermo Fisher Scientific, Cat#34094) were used according to the manufacturerâs instructions. The ECL signal was visualized using Bio-rad ChemiDoc XRS+ and quantified using the Image Lab software (Bio-rad, Version 6.0.1). Each of the experiments was repeated in three independent biological replicates and one representative was shown in the figure.
HERC4 co-immunoprecipitations were performed using the DynabeadsTM Co-Immunoprecipitation Kit (Thermo Fisher Scientific, Cat#14321D) according to the manufacturerâs protocol followed by a western blot (described above). About 50âµl of HERC4 antibody (Abcam, Cat# ab856732) conjugated with 10âmg DynabeadsTM (Thermo Fisher Scientific, Cat#14301), and for each pulldown sample, 1.5âmg of antibody conjugated DynabeadsTM were used with 50âµg of protein. The loading concentration of samples in western blots followed by co-immunoprecipitation was 50âµg protein/well in 20âµl. As a control, an equal amount of input or unbound fraction is run with pulldown samples. Each of the experiments was repeated in three independent biological replicates and one representative was shown in the figure.
Co-immunoprecipitations for ubiquitin pulldowns were performed using the Ubiqapture-Q kit (Enzo Life Sciences, Cat#BML-UW8995) according to the manufacturerâs protocol. To detect captured ubiquitin levels, instead of the provided antibody from the kit, the UBIQUITIN antibody (1:500, CST, Cat#3936) was used. About 50âµg of protein was used to pulldown. As a control, equal amount of input or unbound fraction run with pulldown samples. Each of the experiment repeated in three independent biological replicates and one representative was shown in the figure.
IRF pathway reporter assay
In order to measure IRF pathway activity, an IRF-Lucia luciferase reporter system containing Dual-THP1-Cas9 cells (Invivogen, Cat#thpd-nfis) were used. IRF pathway reporter assays were performed in 96-well plates (TPP, Cat#ZZ707902) where Dual-THP1-Cas9 cell line derivative cells were seeded at a cell density of 0.4 million cells/well and stimulated with 30âµM cGAMP (Biolog, Cat#C161) immediately. After 3âh stimulation, cells were treated with the indicated compounds, and the luciferase measurement was performed 16-h after compound incubation. To detect luciferase activity, QUANTI-Luc⢠(Invivogen, Cat#rep-qlc4r1) was prepared according to the instructions on the datasheet. The reagent (50âµl) was added to a black with clear bottom 96-well plates (Greiner, Cat#655090) where the bottom of the plate was sealed (PerkinElmer, Cat#6005199). About 20âµl of the Dual-THP1 cells from each condition pipetted onto the luminescence reagent in a 96-well plate and luminescence was measured for 0.1âs with the EnVision® Multimode plate reader (Revvity, workstation version 1.14.3049.1193). Each of the experiment repeated in three independent biological replicates. Fold increase in luminescence was calculated by dividing each read to its matching control group (DMSO treated) and results were plotted using Graphpad Prism 9.
HTRF assay
For the HTRF assay, wildtype, Ctrl, or HERC4 sgRNA transduced Dual-THP1 were seeded in 200,000 cells/well in V-bottom 96-wells (Corning, Cat#3894). Cells were stimulated with 30âµM cGAMP (Biolog, Cat#C161) right after seeding. After 3âh stimulation, cells were treated with indicated doses of AK59, QK50, or DMSO control. After 16âh compound treatment, cells were spin down at 1200 rpm for 3âmin and supernatant was taken to proceed with HTRF assay. CXCL10 and IFN-beta cytokine measurements from cell supernatants were performed by HTRF CXCL10 kit (Cisbio, Cat#62HCX10PEG) and HTRF IFN-beta kit (Cisbio, Cat#61HIFNBPEG) according to manufacturerâs instructions. Briefly, standards for each assay are prepared fresh according to the indicated concentrations. For each measurement, pipetted in triple technical replicates in 384-well white low-volume plates (Greiner, Cat#784075). In order to avoid split over of fluorescence, samples were loaded with one well gap in between. Each supernatant sample was pipetted in triplicate technical replicates. The premix of antibodies for each assay was prepared according to the instructions and added on top of standards as well as samples. About 665 and 620ânm measurements were done using an EnVision® Multimode plate reader (Revvity, workstation version 1.14.3049.1193). Delta ratio and %CV are calculated according to the formula provided by the manufacturer. Standard curves were plotted for each assay, and unknown sample measurements were interpolated from the standard cure using GraphPad Prism 9 software. Each assay was performed in three biological replicates, and results were plotted using GraphPad Prism 9 software.
PBMCs isolation from whole blood
Anonymized whole blood was collected under informed consent from healthy volunteers through the Interregionale Blutspende (IRB) of the Swiss Red Cross (SRK) in Bern, Switzerland. 2x 9âml of whole blood taken from four independent donors. Independent donors were used as biological replicates of the data. About 18âml blood diluted with 18âml 1x DPBS (Thermo Fisher Scientific, Cat#14190094) pipetted slowly in SepMate tubes (Stemcell Technologies, Cat#15460) which were already balanced with 15âml Lymphoprep solution (Axis-Shield, Cat#1114547). Samples were centrifuged at 1200âÃâg for 15 min with slow brake, and then the top and middle layers were carefully separated. The platelets were removed with a centrifuge at 114Ãg for 10âmin. red blood cells were removed by RBC lysis buffer (Thermo Fisher Scientific, Cat#00-4333-57). Before seeding for experiments, cells were strained with a 70-µm cell strainer (BD, Cat#352350). For each compound condition, 1 million cells/well in 24-well plates (TPP, Cat#Z707791) were seeded. Samples were treated with either 10âµM of AK59, QK50, or DMSO for 16âh. For STING quantification on human PBMCs after AK59 treatment, cells were treated with 1:50 diluted Fc block (BD, Cat#301804) for 10âmin at room temperature. After the block, cells were stained with CD14 antibody (Biolegend, Cat#325611) in 1:50 dilution for 30 min at room temperature. Wash the cells using FACS Wash Buffer containing 1x DPBS (Thermo Fisher Scientific, Cat#14190094) and proceed with the fixation step of the âSTING quantification by flow cytometryâ protocol.
For analysis, after cells gated for single cells by forward and side-scatter, CD14+ cells were gated for monocytic population in PBMCs and gated group of cells were then further analyzed for STING expression.
STING quantification by flow cytometry
THP1-Cas9 cells were seeded at a density of 1âÃâ106 cells/ml in 24-well plates with the media containing the indicated compound. Compound incubation time varied between 5â16âh and indicated concentrations of bortezomib treatments were always 1âh prior to any additional compound treatment. Cells were always seeded together with the initial treatment. After the compound incubation, cells were collected and fixed using 2.5% paraformaldehyde (stock 32%, Electron Microscopy Sciences, Cat#15714-S) at 37â°C for 10âmin. Cells were then washed using FACS Wash Buffer containing 1x DPBS (Thermo Fisher Scientific, Cat#14190094)â+â0.5% FBSâ+â2âmM EDTA and permeabilized at room temperature for 20âmin using 100âµl of Perm/Wash I (BD, Cat# 557885), diluted 1:10 with 1x DPBS (Thermo Fisher Scientific, Cat#14190094). Cell washing was performed again and then samples were stained with 150âµl/sample anti-STING Alexa488; 1:200 (Abcam, Cat# ab198950) diluted in Robosep Buffer, which contained PBSâ+â2.0% FBSâ+â1âmM EDTA (Stemcell; # 20104) at 4â°C for 1â2âh. After antibody incubation, cells were washed with Wash Buffer three times; then cell pellets were resuspended in FACS Wash Buffer, and flow cytometry acquisition on Fortessa was then performed using Diva software (BD, version 9.0.1). Each of the experiment repeated in three independent biological replicates. Analysis was performed using FlowJo software (version 10.6.1).
Visualization and representation of STING crystal structure
STING structure published by ref. 53 was used, accessed from the PDB database (Entry: 7SII). STING structure was rendered in the PyMOL Molecular Graphics System (version 3.0 Schrödinger, LLC). SAVI mutation information was taken from previous reports36.
STING-GFP construct design and tracking with live cell imaging
Either full-length or cytosolic domains for STING (consisting of either the C-terminal 141â341aa or 155-341aa) were cloned into the pcDNA3.1(+) vector backbone with and without a C-terminal GFP tag. HEK293T cells were seeded on six-well plates (TPP, Cat#Z707759) with a cell density of 1 million cells/well. For live tracking of the protein expression, 1 day after seeding, cells were transfected with the GFP-tagged STING expression constructs using the Trans-IT transfection reagent (Mirus, Cat#MIR2304). After transfection, cells were placed into an Incucyte® (Sartorius, version 2022B rev2) live cell imager where 10x phase contrast and GFP images were taken. Expression of GFP-tagged constructs were observed over 48-h from the start of the transfection until treatment. A total of 16 images from each well were recorded for better coverage, in 2-h time intervals. GFP+ cell area normalized to total cell area per well. The GFP channel was set to identify GFP+ in the cut-offs of minimum 0 and maximum 1000 as a default setting where the background is subtracted.
After 48-h transfection, cells were treated with either DMSO or 10âµM AK59 for 16âh. During treatment the time interval decreased 30â45âmin. Data normalized to the start of the treatment and plotted as the delta GFP (treatment-control). For the HERC4 knockout and Ctrl sgRNA comparison, all the wells were treated with the AK59, and the GFP signal difference between STING155-341 and STING141-341 was measured. Data normalized to the start of the treatment and plotted as the delta GFP (STING141-341– STING155-341). Each of the experiment repeated in three to four independent biological replicates.
For each STING-GFP live imaging experiment, complementary western blot analysis was performed on HEK293T cells transiently transfected with non-GFP-tagged STING constructs in order to decrease background in blots. Each of the experiment was repeated in three independent biological replicates and one representative was shown in the figure.
CRISPR genome-wide screening
For genome-wide CRISPR knockout screening, THP1 cells constitutively expressing Cas9 were generated by lentiviral delivery of the Cas9 protein gene in pNGx-LV-c004 and selected with 5âµg/ml blasticidin S HCl (Thermo Fisher Scientific, Cat#A1113902) as previously described54. For screening, we used a sgRNA library targeting 18,360 protein-coding genes with five sgRNA/gene55. The sgRNA library was packaged into lentiviral particles using HEK293T cells as previously described55,56. Briefly, 2.1âÃâ107 HEK293T cells were seeded into CellSTACK (Corning, Cat#3391) cell culture chambers and transfected with the sgRNA library plasmid mix together with ready-to-use lentiviral packaging plasmid mix (Cellecta, Cat#CPCP-K2A) 24âh after seeding using the Trans-IT transfection reagent (Mirus, Cat#MIR2304). Viral particles were harvested 72âh post-transfection and quantified using the Lenti-X qPCR kit (Clonetech, Cat#631235).
THP1-Cas9 cells were expanded for library transduction. On day zero, the cells were seeded and transfected to achieve a coverage of the library of at least 1000 cells/sgRNA with a multiplicity of infection (MOI) of 0.5. 5âµg/ml polybrene (Millipore, Cat#TR-1003-G) was used in transfection of THP1-Cas9 cells. Transduced cells were selected with 4âµg/ml puromycin for 3 days and on the 4th day, cells were analyzed for RFP expression using the FACS Aria (BD) for determining transduction efficiency. After collecting the day-4 samples, the rest of the library-transduced cells were seeded for the screen with two biological replicates per condition. At day 10, cells were treated with either DMSO or 10âµM AK59 and incubated for 16âh. After compound incubation, cells were harvested, fixed, and stained for STING expression (described in detail below). Then the fraction of cells with the 25% highest and 25% lowest levels of staining for STING, in each treatment group, the sorted samples were processed for genomic DNA isolation using the QIAamp DNA blood maxi kit (Qiagen, Cat#51192) according to the manufacturerâs protocol. Genomic DNA was quantified using the Quant-iT PicoGreen assay (Thermo Fisher Scientific, Cat#P7589) according to the manufacturerâs recommendations and proceeded with Illumina sequencing.
Illumina sequencing of the library
The integrated sgRNA sequences were PCR amplified using primers specific to the integrated lentiviral vector sequence and sequenced using the Illumina sequencing technology. Illumina library construction was performed as previously described55. Briefly, a total of 96âµg of DNA per sample was split into 24 PCR reactions, each with a volume of 100âµl, containing a final concentration of 0.5âµM of each of the following primers (Integrated DNA Technologies, 5644 5â²-AATGATACGGCGACCACCGAGATCTACACTCGATTTCTTGGCTTTATATATCTTGTGGAAAGGA-3â² and INDEX 5â²-CAAGCAGAAGACGGCATACGAGATXXXXXXXXXXGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC-3â², where the Xs denote a ten base PCR-sample specific barcode used for data demultiplexing following sequencing), 0.5âmM dNTPs (Clontech, Cat#4030), 1x Titanium Taq DNA polymerase, and buffer (Clontech, Cat#639242). PCR cycling conditions were as follows: 1âÃâ98â°C for 5âmin; 28âÃâ95â°C for 15âs, 65â°C for 15âs, 72â°C for 30âs; 1âÃâ72â°C for 5âmin. PCR samples were purified using 1.8x SPRI AMPure XL beads (Beckman Coulter, Cat#A63882) according to the manufacturerâs recommended protocol and the qPCR quantified using primers specific to the Illumina sequences according to the sequencing library qPCR quantification guide (Illumina, Cat#SY-930-1010). Amplified libraries were then pooled and sequenced with HiSeq 2500 instrument (Illumina) with 1x 30b reads, using a custom read 1 sequencing primer: 5645 (5â²-TCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCG-3â²), and a 1x 11b index read, using the standard Illumina indexing primer (5â²- GATCGGAAGAGCACACGTCTGAACTCCAGTCAC-3â²), according to the manufacturerâs recommendations.
Analysis of the CRISPR screen
Sequencing analysis was performed as previously described55. In short, raw sequencing reads were converted to FASTQ format using bcl2fastq2 (version 2.17.1.14, retrieved from http://support.illumina.com/downloads/bcl2fastq-conversion-software-v217.html), trimmed to the guide sequence with the fastx-toolkit (version 0.0.13, retrieved from http://hannonlab.cshl.edu/fastx_toolkit/index.html) and aligned to the sgRNA sequences in the library using bowtie57 with no mismatches allowed. Differential presentation of sgRNAs was calculated using DESeq258 and gene-level results were obtained using the redundant siRNA activity (RSA) algorithm59. In RSA, the rank distribution of individual sgRNAs is examined to calculate a hypergeometric enrichment score for the concerted action of each geneâs set of guides. This results in a gene-level p-value for significance. Furthermore, the lower (Q1) or upper (Q3) quartile of the sgRNAâs fold changes to represent effect size at the gene level.
Due to the pooled analysis approach in the RSA algorithm, the range of RSA and Q values in each comparison is in a similar magnitude which enabled further comparisons. In order to rank the hits, RSA and Q values from two different comparisons (in this case, DMSO high vs. low STING and AK59 high vs. low STING) were taken as point coordinates. The Euclidean distance (vectoral) between two coordinates (a.k.a RSA and Q values of each comparison) were then calculated as the magnitude of the vector and the direction of the vector was described by the increase/decrease of RSA and Q values between two points (Bioconductor 4.0.2, Supplementary Data 4). Then, hits were ranked according to the magnitude and the direction of their vector.
STRING protein association analysis
Interaction between STING, HERC4, UBA5, and UBA6 was shown using STRING db. (Version 11.5 with the minimum required interaction score of âmedium confidence (0.4)â or version 12 with the minimum required interaction score of âlow confidence (0.15)â)38.
Individual CRISPR knockouts and TIDE analysis
THP1-Cas9, Dual-THP1-Cas940, and HEK293-JumpIN-Cas9 cells were generated by lentiviral delivery of the Cas9 protein gene in pNGx-LV-c00454 and selected with 5âµg/ml blasticidin S HCl (Thermo Fisher Scientific, Cat#A1113902). Individual knockouts were generated by the lentiviral delivery of sgRNAs in the pNGx-LV-g003 backbone. SgRNA transduced cells were selected with 3.5âµg/ml puromycin (Thermo Fisher Scientific, Cat#A1113802). sgRNA sequences targeting the gene of interests are listed below. Knockout efficiency was checked after the third passage by both Tracking of Indels by Decomposition (TIDE) analysis39 and Western blot.
To check the rate of Indel formation at the CRISPR cut-site, TIDE analysis39 was used as previously described60. Briefly, genomic DNA was extracted from approximately one million cells per condition using a DNA extraction kit (Qiagen, Cat#69504) according to the manufacturerâs recommendations. DNA concentration was measured with the Nanodrop (Thermo Fisher Scientific), and PCR reactions were performed with 2x Phusion polymerase master mix (Thermo Fisher Scientific, Cat#F548S), 5% DMSO, 100âng of DNA and a final concentration of 1âµM forward and reverse primer. PCR primers for each gene are given in Supplementary Table 1. The amplification run was as follows: initial denaturation at 98â°C 30âs, 30 cycles of denaturation at 98â°C for 5âs, annealing at 61â°C for 10âs, extension at 72â°C for 15âs and final extension at 72â°C for 2âmin. PCR samples were cleaned up using PCR and Gel extraction kit (Qiagen, Cat#28704) according to the manufacturerâs protocol and Sanger sequenced. Frameshift, in frame and wildtype calculated from the TIDE analysis results and undefined sequencing reads were excluded.
NanoBiT® complementation assay
Full-length human HERC4 (CCDS41533) was cloned into the pFN35K SmBiT TK-neo Flexi® Vector, encoding an N-terminal small bit (SmBiT) tagged construct (Promega) with the native HSV-TK promoter swapped out for a CMV promoter. C1025A mutation was introduced with site-directed mutagenesis. The cytosolic domain of human STING (either 141â341, 155â341, or 141â341_K150R) was cloned into the pFC34K LgBiT TK-neo Flexi® Vector, encoding a C-terminal large bit (LgBiT) tagged construct (Promega) which is transcribed under the control of the HSV-TK promoter.
Next, 4âÃâ104 HEK293T cells were seeded into 96-well plates (TPP, Cat#ZZ707902). On the same day, cells were transiently transfected with both constructs using a Trans-IT transfection reagent (Mirus, Cat#MIR2304) according to the manufacturerâs recommended protocol. Then 48âh after transfection, cells were treated with the indicated doses and incubation times of either DMSO, AK59, or QK50. For the dose-response, all samples were measured at 16-h post-compound treatment. For the QK50-AK59 competition, QK50 (or DMSO control) treatment started 3âh prior to 16-h AK59 treatment with the indicated doses. For the cGAMP competition with AK59, cGAMP stimulation was initiated by the addition of 30âµM of cGAMP three hours prior to the 16-h compound treatment. To detect the luciferase activity, the Nano-Glo® luciferase assay system (Promega, Cat#N1110) was used according to the manufacturerâs protocol and luminescence was measured for 0.1âs with EnVision® Multimode plate reader (Revvity, workstation version 1.14.3049.1193). Each of the experiment repeated in three to four independent biological replicates. Fold increase in luminescence was calculated by dividing each read to its matching control group (DMSO treated) and results were plotted using Graphpad Prism 9.
Statistics
Statistical analysis of the indicated data was performed using GraphPad Prism 9. Data points were represented as the meanâ±âstandard deviation (SD). Appropriate statistical tests for each data were indicated in the figure legends. Significance indicated as ns pâ>â0.05, *pâ<â0.05, **pâ<â0.01, ***pâ<â0.001, ****pâ<â0.0001. Additionally, exact p values were indicated wherever it is possible.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.