Architecture of the CaV1.2 channel complex
To investigate the inhibitory mechanism of CaV1.2-targeting drugs, we co-expressed the human CaV1.2 with its physiological auxiliary subunits α2δ1 and β2b in HEK293 cells. The genes encoding these three subunits were subcloned into pEG-BacMam vector. We also carried out whole-cell patch clamp using HEK293T cells to characterize the functional properties of these constructs (Supplementary Fig. 1a). The subunits expressed with these constructs form a functional complex, showing an activation curve similar to those from previous studies31,32, and the half-activation voltage (V1/2) of the recombinant channel is +1.0 (±0.5) mV. Subsequent purification experiments resulted in a sharp and symmetrical SEC profile, indicating homogeneity of the sample of the channel complex. The SDS-PAGE gel confirmed that the purified sample contained all three subunits (Supplementary Fig. 1b, c). Next, we collected cryo-EM data using Titan2 Krios and carried out single-particle cryo-EM analysis. The three-dimensional cryo-EM maps of CaV1.2 in the apo state (denoted as CaV1.2apo), tetrandrine bound state (CaV1.2TET), and benidipine bound state (CaV1.2BEN) were determined at 3.5-à , 3.4-à , and 3.3-à resolutions, respectively. These maps are rich in structural features, including densities for sidechains, N-glycans, and lipid molecules, and allowed us to reliably build atomic models of these CaV1.2 complexes (Supplementary Figs. 2â3) (Supplementary Table 1).
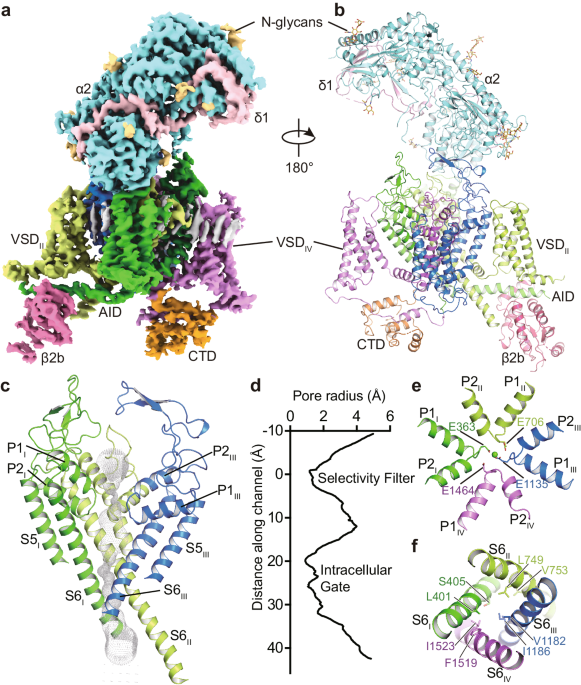
The structure of the CaV1.2 channel complex is composed of a pore-forming α1 subunit and auxiliary subunits α2δ1 and β2b (Fig. 1a, b). Its α1 subunit consists of four repeat transmembrane domains (DIâDIV). Each domain contains six transmembrane helices (S1âS6) and forms the channel in a domain-swapped fashion. The S1âS4 helices constitute the voltage sensing domain (VSD) to detect changes in the cross-membrane electrostatic potential. Consistent with all known structures of voltage-gated channels, the S4 helix assumes a 310-helical conformation. The S5âS6 regions of all four domains form the pore domain to facilitate and control the passage of ions (Fig. 1c, d). The selectivity filter (SF) is contributed by the re-entrant loops from each subunit that connect S5 and S6 and include P1 and P2 helices (Fig. 1e). In contrast to the conformational heterogeneity of the α-interaction domain (AID) found in CaV1.1 (ref. 33), AID is well resolved in the current CaV1.2 structure, with its N- and C-termini adjacent to the intracellular gate and VSDII, respectively (Fig. 1a). The selectivity filter contains four acidic residues (E363DI, E706DII, E1135DIII, and E1464DIV; i.e. the EEEE motif), one from each domain, producing a strongly negatively charged zone to attract cations and determine selectivity for Ca2+ ions33 (Fig. 1e). On the cytoplasmic side, the four S6 helices bundle together and act as a gate to control the access of ions into the pore (Fig. 1f). The pore diameter profile indicates that the current structure represents an inactivated state with a closed intracellular gate (Fig. 1d). Although all S4 helices in the four VSDs are in an âupâ conformation, the intracellular gate remains closed, suggesting that the structure of CaV1.2 was captured in an inactivated state. In addition, the α2δ1 subunit was clearly resolved and sits on the extracellular side of CaV1.2, interacting with the E149, D150, and D151 residues from the DI repeat. The β2b subunit is composed of a GK domain and an SH3 domain and is associated with the cytosolic AID region of CaV1.2 (Fig. 1). A lipid molecule was observed in the DIâDII fenestration of CaV1.2apo, consistent with other structure studies on CaV1.2 (refs. 34,35,36).
a, b The overall EM density map and structure of the CaV1.2 complex. The α2δ1 and β2b subunits, voltage-sensing domain (VSD), C-terminal domain (CTD), α-interacting domain (AID), and N-glycans were labeled. The α1 subunit is colored in deep green (Domain I, DI: A114-R211, F231-I447), light green (DII: W509-K778), deep blue (DIII: F889-I943, F964-Y1197), mauve (DIV: K1198-S1321, E1350-T1357, S1372-E1484, P1494-D1533), and orange (CTD: W1534-P1574, C1582-S1593, N1607-K1638, K1647-P1655), respectively. The β2b (D222-R276, L297-W407), α2 (F27-D129, Q143-V224, K232-N540, Q554-K690, N698-T828, D849-P906), and δ1 (Q972-G1022, T1028-D1070) subunits are colored in magenta, turquoise, and pink, respectively. N-glycans are colored in yellow. c The ion permeation path in the pore domain. The selectivity filter and S5-S6 helices are shown in cartoon and viewed in parallel to the membrane plane. The ion conducting pathway was calculated by using the program HOLE and illustrated with gray dots. d Plot of pore radii for the CaV1.2 complex. e The selectivity filter ring of four glutamate residues from the four domains of CaV channel were shown in sticks. A cation ion is shown as a green sphere. f The intracellular gate formed by four S6 helices viewed from the intracellular side. Hydrophobic residues are shown in sticks.
The structure of CaV1.2apo was compared with that of CaV1.1 (PDB ID: 5GJW)33, giving rise to an RMSD of 1.35âà for 1772 Cα-pairs. The overall structure, including VSD, SF, and extracellular loops (ECLs), is fairly superimposable (Supplementary Fig. 4a). In both structures, in the absence of applied membrane potential, all VSDs show an âupâ conformation of the S4 helices and are thus in the âactivatedâ state. However, some structural differences are observed between the structures of CaV1.2apo and CaV1.1. For example, two Ï-bulges occur on the S6DI and S6DIII helices of CaV1.2apo, whereas both helices assume a canonical α-helix conformation in CaV1.1. The sidechains of residues on the intracellular segment, starting from F394S6I and F1175S6III, undergo an approximate 90° rotation (Supplementary Fig. 4b). The residues forming the central cavity and the intracellular gate differs from that in CaV1.1. In specific, the side chain of F394 and F1175 face the central cavity, causing a shrinkage of the inner radius of the pore domain (Supplementary Fig. 4c, d). In CaV1.1, the intracellular gate is constituted by V329, L333, F656, A660, F1060, V1064 F1376, and I1380. With the local conformational shifts on the S6 helices in CaV1.2, the intracellular gate is formed by L401, S405, L749, V753, V1182, I1186, F1519, and I1523 (Supplementary Fig. 4e). A previous study on TRPV6 channel suggested that the transition from an α-helix to a Ï-helix may prompt channel opening37. However, despite such transition being observed in CaV1.2, its intracellular gate remains closed (Supplementary Fig. 4f).
Pathogenic mutations of CaV1.2
Dysfunction of CaV1.2 usually causes heart and vessel diseases and neurological disorders18,19,20. At least 86 mutations of the CaV1.2 α1 subunit have been identified in various cases of human diseases, including Timothy syndrome (TS), long QT syndrome 8 (LQTS), short QT syndrome (SQTS), Brugada syndrome (BrS), autism, and schizophrenia, among others (Supplementary data). Our current CaV1.2 structure provides a template to map these disease-related mutations into the 3D structure. Among these mutants, 39 sites are distributed on the four VSDs, pore domain, and C-terminal domain (CTD) (Fig. 2). The VSDII and pore domain harbor most of the pathogenic mutations. The remaining 47 mutation sites are mainly distributed on the N-terminus, cytosolic long linkers between different domains (mainly DIIâDIII), and the C-terminal tail, and they were not determined due to conformational heterogeneity.
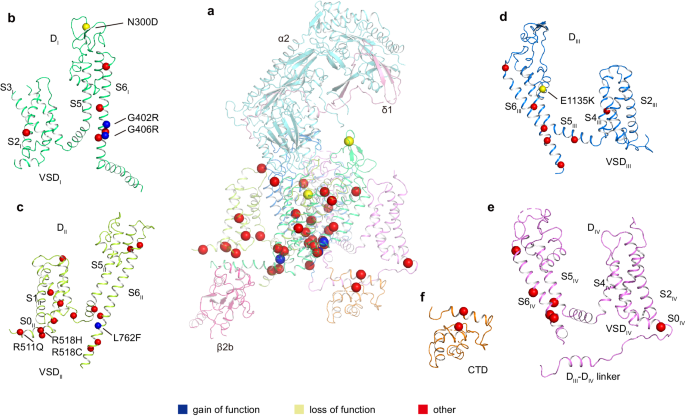
aâf Disease related mutations are located on DI (b), DII (c), DIII (d), DIV (e), and CTD (f). DI, DII, DIII, DIV, and CTD of α1 subunit are colored in deep green, light green, deep blue, mauve, and orange, respectively. Subunits α2, δ1, and β2b are colored in turquoise, pink, and magenta, respectively. Mutation sites are marked with spheres and colored in blue (gain of function), yellow (loss of function), and red (other), respectively.
Three gain-of-function (GOF) mutations have been identified and verified by electrophysiological experiments. G402S and G406R were found in Timothy syndrome cases, and they inhibit voltage-dependent inactivation of CaV1.2 (refs. 18,38). Since all G406 substitutions to other amino acid residues, such as alanine, serine, and valine, impair the inactivation process, the function of G406 is proposed as irreplaceable38. In our CaV1.2 structure, both G402 and G406 are located on the S6DI helix and close to the intracellular gate (Supplementary Fig. 5b). We speculate that the absence of a sidechain in glycine at these two positions is essential for the conformational transition from the open state to the inactivated state, and replacement by a sidechain-containing residue at either of these two positions probably hinders the transition.
In a previous study, L762F associated with LQTS was found to result in a GOF defect with slower inactivation39. In the current CaV1.2 structure, L762 on the S6DII helix is located around the cytosolic membrane surface. Its sidechain points toward the AID motif and the S4-S5DII linker helix (Supplementary Fig. 5c). We speculate that the mutation L762F results in steric hindrance with AID or S4-S5DII helices, leading to a conformational change in these helices and affecting channel inactivation kinetics.
N300D is a loss-of-function (LOF) mutation and has been identified in Brugada syndrome patients40. This mutation leads to a decrease in the current density by impairing the expression level of CaV1.2 in the membrane40. N300 is located in the extracellular loop between S5DI and S6DI (Supplementary Fig. 5a) and is exposed to the interface between the α1 and α2δ1 subunits, which is essential for trafficking of the CaV1.2 complex41,42. N300D may thus affect traffic by disturbing the interaction with α2δ1. In addition, E1135K is another LOF mutation identified in patients with idiopathic QT prolongation, bradycardia, and autism spectrum disorder43,44. This mutation is positioned in the EEEE motif of the selectivity filter in the current CaV1.2 structure (Fig. 1e and Supplementary Fig. 5d). Electrophysiological experiments demonstrated that the E1135K mutation makes CaV1.2 a nonselective monovalent cation channel, in line with functional roles of the EEEE motif in determining ion selectivity44,45.
Previous studies have indicated that R511Q, R518C, and R518H are associated with LQTS and cardiac only Timothy syndrome46,47. In the current CaV1.2 structure, R511 and R518 are located at a cytosolic amphiphilic helix N-terminal to S1DII (denoted as S0DII). The S0DII helix is rich in positive charged residues (511RFCRRKCRAAVK522) that electrostatically interact with the negatively charged C-terminus of AID (Supplementary Fig. 5e). Electrophysiological experiments demonstrated that pathogenic mutations associated with R511 and R518 result in slower inactivation46,47. We speculate that these mutations interfere with the interaction between S0DII and AID and thus affect the conformation of AID relative to the pore domain, resulting in reduced inactivation. This speculation is consistent with the notion that AID is important for regulating channel inactivation48,49.
Recognition of tetrandrine
Tetrandrine is a traditional Chinese clinical agent for autoimmune disorders, cardiovascular diseases, and hypertension. It is extracted from the root of Stephania tetrandra S Moore and inhibits both L-type and T-type CaV channels25,50,51. To explore the molecular basis how tetrandrine blocks the activity of CaV1.2, tetrandrine (10âμM) was added during protein expression and purification. We determined the tetrandrine-bound CaV1.2 complex (CaV1.2TET) at 3.4-à resolution (Supplementary Fig. 2). In our cryo-EM map, we found a well-resolved triangle-shaped density in the central cavity of the pore domain, which is well fitted with the tetrandrine molecule and positioned proximal to the selectivity filter (Fig. 3aâc). The two major moieties of the 6,7-dimethoxy-2-methyl-1-benzyl-isoquinoline ring are positioned close to the DIIâDIII fenestration site and DIV, and they are stabilized by forming hydrophobic or Van Der Waals interactions with surrounding residues from the selectivity filter and S6 helices (Fig. 3d, e). In particular, the residues F1175S6III, M1178S6III, A1512S6IV, I1516S6IV, and F1519S6IV play important roles in stabilizing tetrandrine. The 6,7-dimethoxy and 2/2â-N groups from the isoquinoline ring form hydrogen bonds with the N741S6II and Y1508S6IV/N1179S6III residues. In addition, in agreement with observations in drug-bound CaV3.3 complex structures52, a lipid molecule penetrates through the fenestration site between DI and DII and participates in hydrophobic interactions with the tetrandrine molecule (Fig. 3d, e). Compared with other drug binding modes observed in the previously reported CaV1.1 structures, we found that the tetrandrine binding mode is distinct from those of nifedipine (a DHP derivative), verapamil (PAA), diltiazem (BTZ), and cinnarizine (CIN) (Supplementary Fig. 6a). In particular, tetrandrine completely squeezes the binding site of BTZ (Supplementary Fig. 6b), consistent with previous experimental results showing that tetrandrine and diltiazem cannot simultaneously bind to L-type CaV channels53. To validate the binding site of tetrandrine, we designed two mutants, N741W and N1179W, with the potential to create steric clash between tetrandrine and the binding site. We determined the dose-response curves of tetrandrine for both wild-type and these two mutants. The curves for both mutants exhibited a rightward shift compared to that of wild-type CaV1.2, indicating that these mutants reduce the sensitivity of these mutants to tetrandrine. The derived IC50 values for tetrandrine with the N741W and N1179W mutants are 71.3ânM and 41.8ânM, respectively, which are higher than that of wild-type (~14.2ânM) (Fig. 3), further confirming the binding site of tetrandrine.
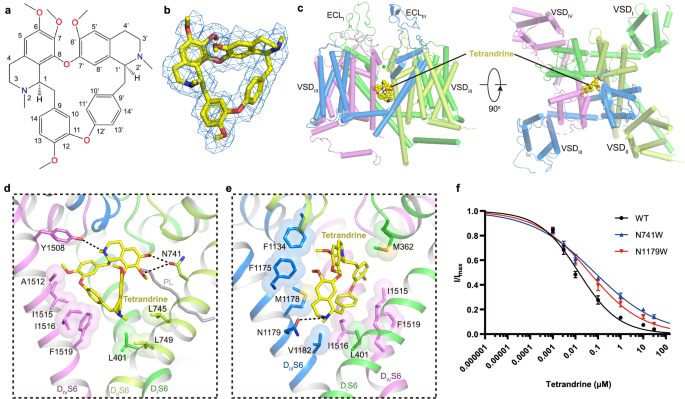
a Chemical structure of tetrandrine. b The cryo-EM density shown in blue mesh for tetrandrine in sticks. c The overall structure of the CaV1.2TET complex. The domains of CaV1.2TET are colored as DI in deep green, DII in light green, DIII in deep blue, and DIV in mauve. The tetrandrine located in the pore domain is presented as yellow spheres. d, e Detailed binding sites for tetrandrine showing interactions between tetrandrine and CaV1.2. The sidechains of key residues are displayed in sticks and the hydrophobic side chains are overlaid with transparent surfaces. Black dashed lines indicate potential hydrogen bonds. A lipid located within the fenestration site is shown in gray. f Dose-response curves of tetrandrine inhibition on CaV1.2WT, CaV1.2N741W, and CaV1.2N1179W. CaV1.2N741W and CaV1.2N1179W shift the IC50 of tetrandrine on CaV1.2 from 14.2ânM to 71.3ânM and 41.8ânM, respectively. Data in curves are represented as meanâ±âSEM (nâ=â10 biologically independent experiments). Source data are provided as a Source Data file.
Antagonism of benidipine
1,4-Dihydropyridine derivatives (DHPs), such as nifedipine, selectively inhibit certain voltage gated calcium channels and are widely used clinically for the treatment of hypertension54. However, DHPs frequently cause side effects such as proteinuria, upon chronic use. The specific inhibition of L-type calcium channels is usually an advantage but is a defect in the kidney. The reason is that L-type channels are distributed in the kidney glomerulus afferent arteriole but are absent in the efferent arteriole. This inconsistent inhibition results in unbalanced local blood pressure, leading to kidney damage with proteinuria55,56. However, benidipine, another DHP derivative, can additionally inhibit T-type calcium channels, which are more abundant in efferent arterioles than in afferent arterioles, making blood pressure more balanced in different parts of the kidney. Compared with nifedipine and amlodipine, benidipine causes fewer adverse side effects, such as proteinuria and leg edema55,57,58; therefore, it is widely used in the clinic. Compared with nifedipine, benidipine replaces 8-nitrophenyl with 9-nitrophenyl and 3-carboxylic acid methyl ester with 3-(1-phenylmethyl-3-piperidinyl) ester. By supplementing benidipine when solubilizing the membrane and before preparing the cryo-EM sample, we obtained a 3.3-à resolution benidipine-bound CaV1.2 structure (CaV1.2BEN) (Supplementary Fig. 2).
The benidipine molecule penetrates into the DIIIâDIV fenestration and corresponds to a âclawâ shape density in the map (Fig. 4a, b). Unlike the pore-blocker tetrandrine, benidipine does not directly block the pore but allosterically inhibits CaV1.2. The main moiety of dihydropyridine is biased towards DIII (Fig. 4c). The nitrogen atom on the pyridine ring forms a potential hydrogen bond with S1132P1 (Fig. 4d). The nitrogen atom on 9-nitrophenyl and the oxygen atom on 5-dicarboxylic acid of the pyridine ring form hydrogen bonds with T1056S5III and Q1060S5III, respectively (Fig. 4d). Whereas similar interactions with T935 and Q939 have been identified in the structure of the CaV1.1 complex, benidipine also contains a phenylmethyl piperidinyl group, which is positioned in a hydrophobic pocket formed by residues I1046S5III, I1049S5III, V1053S5III, F1181S6III, M1509S6IV, and F1513S6IV, providing additional hydrophobic interactions to stabilize its binding, in line with the inhibitory IC50 of benidipine being approximately ten times smaller than that of nifedipine59 (Fig. 4d, e). Previous studies indicated that nifedipine specifically inhibits L-type CaV channels, showing no sensitivity to N- and T-type CaV channels60,61,62. In contrast, benidipine, in addition to L-type CaV channel, can also inhibit both N-type and T-type CaV channels, with IC50 values of approximately 35âμM and 11âμM, respectively59,63,64. To understand the molecular basis of how benidipine is recognized by CaV3.1, we compared the benidipine-bound CaV1.2 structure with that of CaV3.1, and the result illustrates that most of the residues participating in benidipine binding are also conserved in CaV3.1, such as V1053S5III, F1129DIIIP1, and F1513S6IV. Although I1046S5III, M1178S6III, F1181S6III, and M1509S6IV in CaV1.2 are substituted by L1386S5III, V1505S6III, M1508S6III, and L1813S6IV in CaV3.1, respectively, we speculate that these substitutions by similar residues are tolerable and that the relatively conserved binding pocket underlies the basis for how benidipine acts as an inhibitor in multiple voltage gated calcium channels (Fig. 4f). Moreover, two residues, T1056 and Q1060, play pivotal roles in the binding of DHP drugs to L-type CaV channels. However, these residues are not conserved in N-type and T-type CaV. Residue T1056 of CaV1.2 is substituted by Y1289 in CaV2.2, and residue Q1060 is replaced by M1293 in CaV2.2 and F1400 in CaV3.1 (Supplementary Fig. 7a, b, Fig. 4f). Although benidipine does not inhibit mutations T1056Y, Q1060M, and Q1060F as potently as it does in WT CaV1.2, it still retains the ability to block the current (Supplementary Fig. 7c). In contrast, previous studies demonstrated that these same substitutions at positions T1056 and Q1060 almost abolished the sensitivity of CaV1.2 to the DHP drug R202-791 (ref. 65). The varied effects of these mutations on benidipine and R202-791 might arise from the phenylmethyl piperidinyl group in benidipine. This group establishes more interactions with the channel and appears to tolerate substitutions at positions T1056 and Q1060, especially in N- and T-type CaV channels.
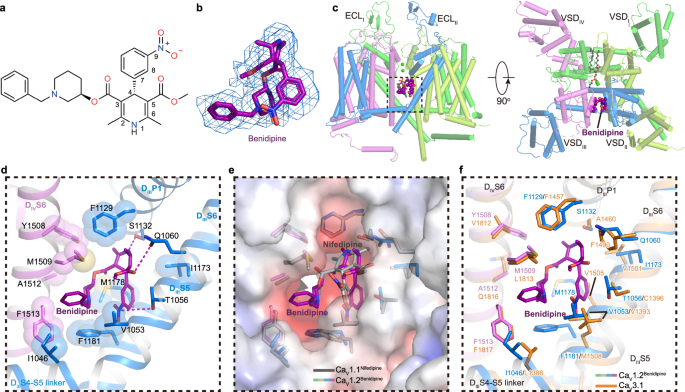
a Chemical structure of benidipine. b The cryo-EM density shown in blue mesh for benidipine in sticks. c The overall structure of the CaV1.2BEN complex. The domains of CaV1.2BEN are colored as DI in deep green, DII in light green, DIII in deep blue, and DIV in mauve. The tetrandrine is presented as violet spheres. Two cation ions are shown as green spheres. Phospholipid entering through fenestration is shown as gray sticks. d Detailed binding sites for benidipine showing interactions between benidipine and CaV1.2. The sidechains of key residues are displayed in sticks and the hydrophobic side chains are overlaid with transparent surfaces. Black dashed lines indicate potential hydrogen bonds. e Comparison of the DHP ligands binding sites of CaV1.2 with nifedipine bound CaV1.1 structure (PDB ID: 6JP5) (colored in gray), overlaid with the electrostatic surface potential of CaV1.2. f Comparison of the benidipine binding sites of CaV1.2 with CaV3.1 (PDB ID: 6KZO) (colored in orange).
Several dihydropyridine drugs binding L-type calcium channels complex structures have been resolved in previous study21,22,66,67,68, including amlodipine bound CaVAb (7KMD), nimodipine bound CaVAb (7KMF), nifedipine bound CaV1.1 (6JP5), amlodipine bound CaV1.1 (7JPX), (S)-Bay K 8644 bound CaV1.1 (6JP8) and (R)-Bay K 8644 bound CaV1.1 (7JPW). When CaV1.2BEN and these structures are superimposed, a same binding pocket in DIII-DIV fenestration are revealed to accommodate benidipine in CaV1.2 and amlodipine/(S)-Bay K 8644/(R)-Bay K 8644 in CaV1.1 (Supplementary Fig. 8aâc). However, benidipine binding site do not overlap with binding sites of amlodipine and nimodipine in CaVAb (Supplementary Fig. 8dâf), and the residues involved in interaction are not conserved as well.
L-type selective dihydropyridine (DHP) inhibitors and L/T-selective DHP inhibitors are cataloged in Supplementary Table 2. The structural similarities among Class 1, 2, and 3 necessitate collective consideration. Upon scrutinizing their chemical structures and inhibitory selectivity, we identified primary distinctions in functional group modifications, specifically additional alterations on the C3 and C4 benzene rings. Within Class 1 and Class 2, with the exception of three instances (cilnidipine, amlodipine, nimodipine), all DHP derivatives predominantly featured smaller, relatively hydrophilic C3 groups for L-type selective compounds, while L/T-type selective compounds displayed larger and hydrophobic C3 groups. For Class 4 and Class 5 compounds, characterized by a modification into a ring at the C5/C6 position, the sole difference between the L-type selective Class 4-1 and L/T-type selective Class 4-4 compounds lies in the presence of a benzoic acid group on the C4 benzene ring in Class 4-1, while Class 4-4 has a hydroxyl group at the corresponding position. Thus, for compounds with C5-C6 rings, the hydrophobic nature of C3 modification does not exert a significant impact; the pivotal factor influencing differences is the presence of an additional aromatic group modification on the C4 benzene ring.
To further investigate interactions among compounds of varied selectivity on L-type and T-type channels, CaV1.2BEN served as a docking template. L-type selective compounds from Class 1 were docked onto L-type channel CaV1.2, while L/T-type selective compounds from Class 1 were docked onto both L-type channel CaV1.2 and T-type channel CaV3.1. Irrespective of L-type or L/T-type selectivity, these compounds bound to the DIII-DIV fenestration in CaV1.2 or CaV3.1, exhibiting a favorable superposition of the dihydropyridine backbone (Supplementary Fig. 9b, c). Both L-type selective and L/T-type selective DHPs docked to CaV1.2 formed hydrogen bonds with T1056, Q1060, and S1132. However, the C3 group of L-type selective DHPs docked to CaV1.2 is confined near M1509 within the hydrophobic pocket formed by DIIIS6 and DIVS6. In contrast, L/T-type selective DHPs docked to CaV3.1, while unable to form hydrogen bonds at the corresponding positions, not only occupy the binding pocket formed by DIIIS6 and DIVS6 but also extend to the hydrophobic pocket formed by DIIIS5 and DIVS5, akin to the position occupied by L/T-type selective DHPs docked to CaV1.2. This suggests that the ability of the C3 group to occupy the binding pocket formed by DIIIS5 and DIVS5 may be pivotal in determining the L-type and L/T-type selectivity of DHPs. In the case of cilnidipine, its C3 benzenyl group occupies the C5 position in the CaV1.2 binding pocket, possibly due to the planar rigidity of the phenyl group preventing it from fitting into the C3 position. The excessively large C5 group may clash with F1400 in CaV3.1, elucidating why cilnidipine, despite its large and hydrophobic C3 group, cannot inhibit T-type channels. In the case of the other exception, amlodipine docked in CaV3.1, the C2 amino group extends towards the pore and forms potential interactions with hydrophilic residues K1462 and Q1868 near the pore, compensating for the modest interaction between the C3 group and CaV3.1. Given that L/T-type selective DHPs also exhibit different affinities for various T-type channels, such as nimodipineâs preference for inhibiting CaV3.2 and CaV3.3 over CaV3.1 (ref. 64), we aligned the CaV3.3 structure with the CaV3.1 structure. The alignment results revealed that the hydrophobic pockets formed by DIIIS5 and DIVS5 in CaV3.3 are smaller than those formed in CaV3.1, potentially explaining why nimodipine produces distinct inhibitory effects on T-type channels with different α1 subunits.