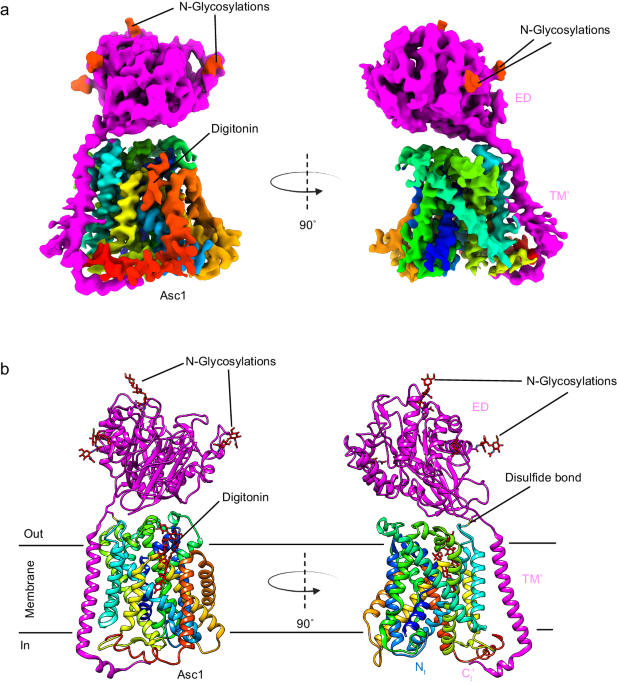
CryoEM of human Asc1/CD98hc
We determined the structure of the human Asc1/CD98hc heterodimer in the absence of substrate in an inward-open conformation with a partial occlusion of TM1a (Fig. 1, Supplementary Table 1), as described in detail below. After expression in HEK293-6E cells, Asc1/CD98hc was purified by NiNTA affinity chromatography and a final step using size exclusion chromatography (Supplementary Fig. 1). The peak fraction of the size exclusion chromatography was applied to holey grids and vitrified, and cryoEM data were collected (Supplementary Fig. 2). Reference-free 2D averages of the extracted particles revealed that most of the ~140âkDa complex was made of well-defined transmembrane helices inserted within the detergent micelle, whereas an ectodomain was placed outside the micelle (Supplementary Fig. 2). We further classified the dataset by using iterative 3D reconstructions and classifications to yield a subset of particles that were refined to high resolution by removing the contribution of noise from the micelle using non-uniform refinement30. The estimated average resolution for the cryoEM map of Asc1/CD98hc was ~ 4âà , with local resolutions as good as 3.4â3.8âà for most of the map, except for some flexible cytoplasmic regions (Supplementary Fig. 2g), thereby allowing the modeling of the Asc1/CD98hc heterodimer structure (Supplementary Fig. 3, 4).
a CryoEM map for Asc1/CD98hc heterodimer. Heavy chain CD98hc is colored in bright pink where the four N-glycosylations have been highlighted in red. Light chain is colored in rainbow from dark blue, blue, cyan, green, yellow, red, from N- to C-terminus. Densities assigned to glycosylations and the digitonin molecule are colored in red and labeled accordingly. The cryoEM map is shown in two different views, which differ approximately 90Ë from each other. b Atomic model of the Asc1/CD98hc heterodimer in the same orientation and color-code than in (a). The disulfide bond that covalently binds CD98hc (C109) to Asc1 (C154) is indicated.
Structure of inward-open semi-occluded Asc1/CD98hc
The cryoEM map for Asc1/CD98hc is in agreement with other reported HATs. It consists of a large density corresponding to the extracellular ectodomain of CD98hc sitting on top of Asc1, whilst Asc1 is mostly formed by transmembrane (TM) helices embedded in the detergent micelle (Fig. 1a). CD98hc contains a TM helix that expands across the membrane and interacts with the cytosolic side of Asc1 (Supplementary Fig. 4a, b). Additionally, the map shows density for the four N-glycosylation sites in CD98hc (Supplementary Fig. 4c). For Asc1, clear density was observed for all 12 TM helices of the transporter, with a lower resolution in TMs 1, 6, 11 and 12, thereby suggesting a certain degree of flexibility in these areas (Supplementary Fig. 2).
The structure of the heterodimer was confidently modeled for residues 41â489 for Asc1 and 61â529 for CD98hc (see Methods for details), thus excluding the flexible N-terminal ends of both proteins (Fig. 1b). Asc1 is covalently bound to CD98hc via a disulfide bridge involving residues Cys 154 (Asc1) and Cys 109 (CD98hc) (Fig. 1b, Supplementary Fig. 4a), with additional contacts between CD98hc TMâ helix and the surrounding TM helices of Asc1. The four N-glycosylation sites of CD98hc (residues 264, 280, 323, and 405) were also modeled (Supplementary Fig. 4c). Finally, density for a digitonin molecule was observed near TM helices 3, 9, 10, and 12 (Fig. 1), as previously reported for human LAT221.
The structure of the Asc1/CD98hc heterodimer has been determined in an inward-open partially occluded conformation where TM1a is displaced towards TM5 when compared to the LAT2/CD98hc structure reported in a fully inward-open conformation (PDB ID 7B00)21 (global rmsd of backbone-only between Asc1 and LAT2 of 1.73âà ) (Fig. 2a, b). In contrast, Asc1/CD98hc is not as closed as shown for the fully occluded conformation of LAT1/CD98hc25 (PDB ID 7DSQ, global rsmd of 3.42âà ) where TM1a and TM5 block the access to the binding cavity from the cytosolic side (Fig. 2b, c). In addition, TM7 in Asc1 is in a position comparable to that of LAT2, thus differing from TM7 in the occluded structure of hLAT1 where TM7 is displaced towards TM1a (Fig. 2d). Therefore, the tilting of TM1a in Asc1 is the main conformational change responsible for partial occlusion of the access to the substrate cavity from the cytosolic side. In addition, we also found differences in a region that connects TM2, TM10, and TM6 in LAT2 and which has been proposed to contribute to maintaining the conformation of TM621. Thus, Asn 249 in TM6b interacts with Tyr 399 in TM10 in the inward-facing conformation of LAT2, whereas Asn 249 is at putative H-bond distance of the side chain of Tyr 93 in TM2 in the partially occluded conformation of human Asc1 (Fig. 2e). This change in the connections established by Asn 249 likely results from the tilting of TM1a observed in Asc1 as the three residues involved are conserved in both LAT2 and Asc1.
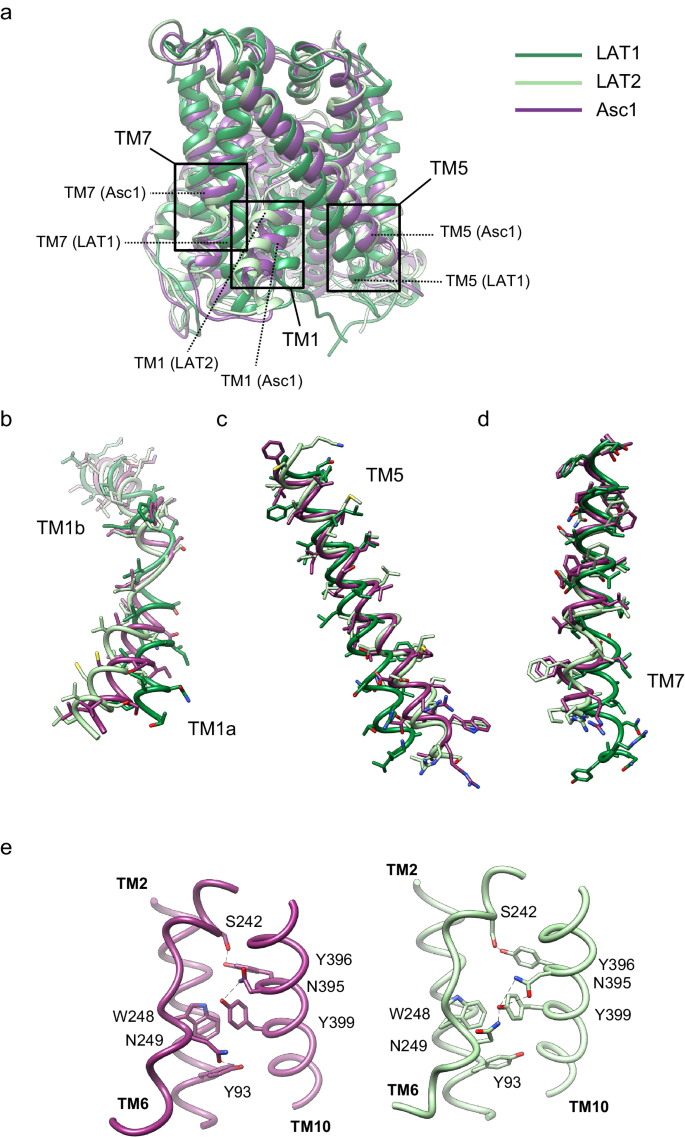
a comparison between the structure of Asc1 (this work) with the structures of LAT1 (PDB ID 7DSQ) and LAT2 (PDB ID 7B00). Models are color-coded as follows: LAT1 in dark green, LAT2 in light green and Asc1 in dark purple. The regions of major differences are highlighted within squares and these regions are enlarged in (bâd). b Details of the differences found in TM1a and TM1b. c Details of the differences found in TM5. d Details of the differences in TM7. e Changes observed in TM10 as scaffold of TM6 between the structures of LAT2 and Asc1.
We then compared the experimental structure of Asc1 with the AlphaFold structural model31,32 (Supplementary Fig. 5a). The prediction matched the overall structural organization of human Asc1 well (with a global rsmd of 1.67âà over the 456 built residues). However, interestingly, discrepancies between the AlphaFold model and the experimental structure were found in regions involved in the function of the transporter: (i) TM1a is less occluded in the structure than in the prediction (Supplementary Fig. 5b) and the position of the backbone of Ile 53 in the C-terminal end of TM1a differs between the experimental structure and the prediction (Supplementary Fig. 5c); (ii) the side chain of Tyr 333 (TM8), a residue that is part of the canonical substrate binding site, is at potential H-bond distance of the side chain of Ser 246 (TM6) in the structure (see later) but the distance between these two residues is much larger in the model predicted by AlphaFold (Supplementary Fig. 5d); and (iii) Lys 194, a key functional lysine residue in TM533 that is conserved in all LATs and the related cationic amino acid transporter GkApcT34, establishes a H-bond with backbone atoms of TM1 in all solved LAT structures in inward-facing conformation19,21,22,23,28 and also in the AlphaFold prediction for Asc1. However, the side chain of residue Lys 194 is not visible in the cryoEM map of Asc1 despite being surrounded by other well-defined residues in TM5 (Supplementary Fig. 5e), thereby suggesting that it does not establish contacts with TM1. This observation correlates well with the fact that the segment of TM1 facing Lys 194 was determined at a lower resolution than surrounding regions of the cryoEM map of Asc1, thus possibly indicating the flexibility of this region in the human Asc1 inward-facing partially occluded structure (Supplementary Fig. 5f).
The Asc1 binding site contains a unique tyrosine in HATs
The substrate binding site of the human Asc1 transporter is in an inward-facing partially occluded conformation, with TM1a tilted to limit the access of substrates to the binding site from the cytosolic side of the membrane. As previously reported for other LATs, the substrate coordination site is formed by the unwound regions of TMs 1 and 619,21,22,23,28, together with other residues in TMs 1a, 6b and 8 (Fig. 3a). In particular, TM1 contains a 55G(S/T)G57 motif with the amide nitrogen atoms of Gly 55, Ser 56 and Gly 57 oriented towards the empty space, providing the possibility of hydrogen bonding with the carboxyl group of substrates. In addition, the unwound segment of TM6 includes Ser 246 with its carbonyl group facing the binding cavity, and Trp 248 facing residues in the adjacent TM2 and TM10 (Fig. 3a). Together, these residues in the unwound regions of TM1 and TM6 generate a cavity with the correct environment for the coordination of the carboxyl and amine groups of substrates.
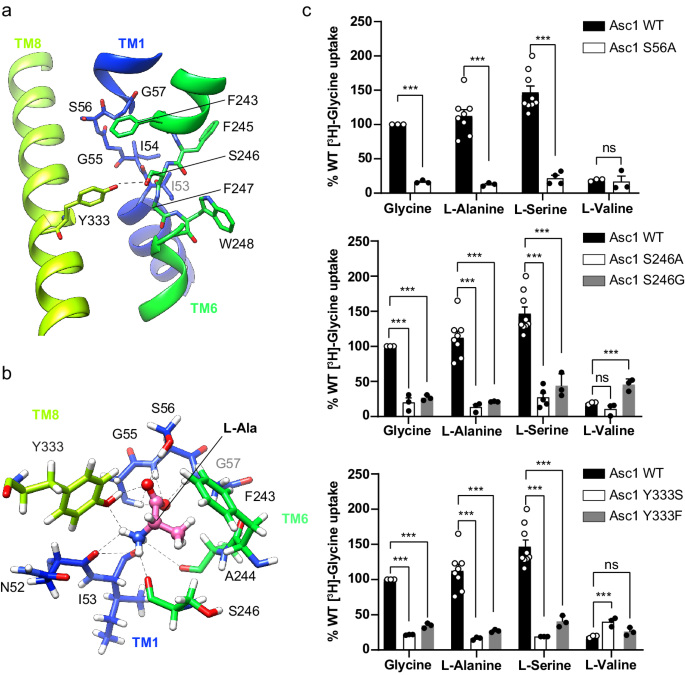
a The unwound segments of TM1 and TM6 plus Phe 243 in TM6 and Tyr 333 in TM8 constitute the substrate binding site. The possible hydrogen bond between Ser 246 and Tyr 333 is indicated with a dashed line. b L-alanine pose 1 in the binding site with the lowest binding energy calculated by PELE. Putative H-bonds are indicated by dashed lines. C atoms in Asc1 residues are colored according with the color of the corresponding TMs as in Fig. 1, substrate C atoms are colored in pink. Another minimum for L-alanine (pose 2) is shown in Supplementary Fig. 8a. c Uptake of the indicated substrates in HeLa cells induced by wild-type (WT) and the indicated Asc1 mutants. Uptake activity is normalized for glycine uptake mediated by WT Asc1/CD98hc (2073â±â223âpmolâmgâ1 protein minâ1 from 17 independent experiments). Data (meanâ±âs.e.m.) corresponds to nâ=â9 (WT L-Ser), nâ=â8 (WT L-Ala), nâ=â5 (S246A L-Ser), nâ=â4 (S56A L-Ser) or nâ=â3 (rest of conditions) independent experiments. P values were obtained by fitting a linear model to Boxcox transformed data and were adjusted for multiple comparisons using the Benjamini-Hochberg method (see materials and methods): ns, non-significant; (***), pâ<â0.001. WT vs S56A Gly, pâ=â7.11âââ10â15; WT vs S56A L-Ala, pâ=â8.88âââ10â16; WT vs S56A L-Ser, pâ<â2.22âââ10â16; WT vs S56A L-Val, pâ=â0,0492; WT vs S246A Gly, pâ=â2.34âââ10â10; WT vs S246A L-Ala, pâ=â1.37âââ10â12; WT vs S246A L-Ser, pâ<â2.22âââ10â16; WT vs S246A L-Val, pâ=â0,0994; WT vs S246G Gly, pâ=â1.11âââ10â14; WT vs S246G L-Ala, pâ=â3.42âââ10â14; WT vs S246G L-Ser, pâ=â1.15âââ10â10; WT vs S246G L-Val, pâ=â1.8910-5; WT vs Y333S Gly, pâ=â4,44âââ10â16; WT vs Y333S L-Ala, pâ=â6.66âââ10â16; WT vs Y333S L-Ser, pâ=â3.11âââ10â15; WT vs Y333S L-Val, pâ=â3.82âââ10â4; WT vs Y333F Gly, pâ=â9.53âââ10â12; WT vs Y333F L-Ala, pâ=â1.92âââ10â12; WT vs Y333F L-Ser, pâ<â1.65âââ10â10; WT vs Y333F L-Val, pâ=â0.153.
In Asc1, the substrate binding site is also complemented by Tyr 333 located in TM8 (Fig. 3a), a residue only present in this transporter within the HATs and found to be important for the mechanism of substrate selectivity and translocation (see below). The hydroxyl groups of Tyr 333 and Ser 246 are at potential H-bond distance, and they delimit one of the edges of the substrate binding cavity (Fig. 3a, Supplementary Fig. 5d).
Interestingly, Asc1 appeared later in evolution than other metazoan LATs, being found only in vertebrates and coinciding with the appearance of Tyr 333 in the substrate binding site of Asc1. Tyr 333 is fully conserved in all available Asc1 sequences (Supplementary Fig. 6). This Tyr residue is also present in BasC, a bacterial LAT transporter with Asc-like transport activity33. This observation thus suggests that this residue contributes to functions in substrate binding and/or transport that are specific to Asc1 among vertebrate LATs, such as the transport of D-serine.
TM1, TM6 and TM8 contribute to Asc1 substrate recognition
Despite the addition of 10âmM D-serine to the purified heterodimer before sample vitrification, we found no density for the ligand in the putative binding site. To overcome this limitation, we performed molecular docking and PELE studies35 using the structure of Asc1/CD98hc and a set of amino acids that are substrates for Asc1 (L-alanine, L-serine, and D-serine). The PELE analysis starts by docking the substrate inside the binding cavity, which then can explore different binding poses, thus overcoming the lack of bound substrate in our structure. This approach predicted minimal binding-energy modes for all the amino acids tested (Supplementary Fig. 7). In the calculated poses, the α-carboxyl of these substrates are predicted to establish H-bonds with the nitrogen atom of residues Gly 55, Ser 56, and Gly 57 in the GSG motif of the unwound segment of TM1, and with the hydroxyl group of Ser 56 in TM1. In contrast, the α-amino of the substrates are predicted to establish H-bonds with carbonyl oxygen atoms of Asn 52 and/or Ile 53 in TM1, as well as with Ser 246 in TM6 (Fig. 3b, Supplementary Fig. 8aâd). Furthermore, each substrate established additional interactions with specific residues in TM6 and TM8, as discussed below.
PELE analysis revealed two minima for L-alanine (Supplementary Fig. 7). In the lowest-energy pose (pose 1), the side chain of Tyr 333 interacted with both the α-carboxyl and α-amino groups of the substrate (Fig. 3b). In the other PELE pose (pose 2), the side chain of Tyr 333 was oriented towards TM6, causing a shift in the interaction network of the substrate, which is then predicted to establish H-bonds with the side chain of Ser 246 (Supplementary Fig. 8a).
In the case of L-serine and D-serine, PELE analysis revealed that the α-amino and α-carboxyl groups of both substrates share similar interaction networks (Supplementary Fig. 8bâd). Nevertheless, some differences were observed in the side-chain hydroxyl interactions. For L-serine, we obtained two poses with similar binding energy values (Supplementary Fig. 7): one where the substrate hydroxyl group interacted with Phe 243 in TM6 (Supplementary Fig. 8c) and another where it interacted with Tyr 333 in TM8 (Supplementary Fig. 8d). D-serine presented a single minimum where the substrate hydroxyl group interacted with Phe 243 (Supplementary Fig. 8b).
Our PELE studies indicated that human Asc1 binds its substrates between the unwound regions of TM1 and TM6 and identified a series of residues in TM1 (Ser 56), TM6 (Ser 246) and TM8 (Tyr 333) whose side chains might participate in substrate recognition. Remarkably, in Asc1 a putative interaction has been observed between the substrate and TM8, thus connecting the bundle (TMs 1, 2, 6, and 7) and scaffold (TMs 3, 4, 8, and 9) domains, a similar design to that previously reported for the binding of L-arginine in the prokaryotic homolog AdiC36.
Ser 56, Ser 246 and Tyr 333 are determinants of selectivity
PELE analysis identified several residues whose side chain interacts with the substrate in the binding site (Ser 56, Ser 246, and Tyr 333). Thus, we performed functional studies of Asc1 mutants predicted to interfere with substrate binding (S56A, S246A, S246G, Y333S, and Y333F). To this end, we measured amino acid uptake in HeLa cells co-transfected with either the human wild-type or the mutated version of Asc1, together with human CD98hc. The expression levels and subcellular localization of these Asc1 mutants were comparable to those of the wild-type transporter (Supplementary Fig. 9). All these mutations caused a dramatic decrease in the uptake of small neutral substrates ([3H] glycine, [3H] L-alanine and [3H] L-serine) but did not affect (in the case of S56A and S246A) or slightly increased (in the case of Y333F, Y333S and S246G) the uptake of the larger amino acid ([3H] L-valine) (Fig. 3c). These results indicate that Ser 56, Ser 246 and Tyr 333 establish interactions with the substrates that are essential for the transport of small amino acids but not for larger ligands (e.g., branched-chain amino acids) such as L-valine, for which Asc1/CD98hc is a much poorer transporter1. L-valine might require the participation of other residues in Asc1.
Asc1 shows distinctive properties in its acceptance of D-amino acids1, although the molecular bases underlying this lack of stereoselectivity have not been yet described. Consequently, the transport of [3H] L- and D-serine in the S56A, S246A, S246G Y333S, and Y333F mutants of Asc1 co-expressed with CD98hc was studied in HeLa cells. We observed a comparable decrease in the uptake of both enantiomers, in agreement with the highly similar interaction networks that these two substrates shared in the PELE binding poses (Supplementary Fig. 8e).
Taken together, our results suggest that the residues studied (Ser 56, Ser 246, and Tyr 333) are essential for the transport of small neutral amino acids and that point mutations of these residues do not alter the selectivity of the transporter towards the enantiomers of serine. We hypothesize that evolution has sculpted the binding site of Asc1 to transport small neutral amino acids in a non-stereoselective manner, with three essential interactors that are shared between serine enantiomers. Having established that these residues are essential for transport activity, we then proceeded to investigate further into their possible mechanistic role in the two transport modes presented by Asc1, namely facilitated diffusion and exchange.
Tyr 333 is important for facilitated diffusion
Asc1/CD98hc is the only HAT of neutral amino acids that presents two distinct transport modes, namely amino acid exchange and facilitated diffusion1,7,8. Considering that Tyr 333 is a residue found only in Asc1 within vertebrate LATs (Supplementary Fig. 6), that the hydroxyl groups of Tyr 333 and Ser 246 are predicted to be at possible H-bond distance in our structure (Fig. 3a, Supplementary Fig. 5d), and that this potential bond would connect the scaffold and bundle domains in an analogous manner to the substrate in our PELE analysis (Fig. 3b, Supplementary Fig. 8aâd), we hypothesized that these residues participate in the facilitated diffusion mode of transport of Asc1. MD analysis of our apo structure predicted that the interaction between the side chains Ser 246 and Tyr 333 is weak, being present in less than 2% of the frames of the simulation (Supplementary Fig. 10). Nevertheless, given that our structure in a partially occluded conformation is likely to represent a transitory state between more stable states and that we cannot rule out a stronger interaction between Tyr 333 and Ser 246 in another conformation (i.e., fully occluded), we performed functional assays to obtain further insight into the possible role of this connection in facilitated diffusion.
To this end, we set up a system to measure and compare amino acid transport by exchange and by facilitated diffusion in HeLa cells overexpressing wild-type Asc1/CD98hc and selected mutants (S246G and Y333F) (Supplementary Fig. 11). The efflux of radioactivity from cells preloaded with [3H] D-serine was measured in linear conditions in the following three conditions (Supplementary Fig. 11aâd): (i) in the presence of the specific Asc1 inhibitor BMS466442 at full-blocking concentration of 5âµM12 to measure efflux of radioactivity not mediated by Asc1/CD98hc (Supplementary Fig. 11b). We consistently detected less than 20% of the loaded radioactivity coming out from inside the cells in the presence of the inhibitor, which was considered as experimental background (Supplementary Fig. 11e, f); (ii) in the absence of amino acid outside the cells (Supplementary Fig. 11c), radioactivity could only be transported by Asc1 from the cytosol to the extracellular space by facilitated diffusion, and these conditions were achieved by performing the experiments in an amino acid-free medium (Supplementary Fig. 11f); (iii) in the presence of extracellular D-serine at saturating concentrations (3âmM; 15â20-fold Km value) (Supplementary Fig. 11d), to measure transport by the exchange mechanism of Asc1 (Supplementary Fig. 11e). In these two latter cases, to estimate the transport by Asc1 corresponding to facilitated diffusion or exchange, we subtracted the background measured in the presence of the Asc1/CD98hc inhibitor performed in parallel from each measurement.
Efflux via Asc1 was expressed as the percentage of the loaded radioactivity released to the medium per minute after subtraction of the background measured in the presence of the inhibitor (Fig. 4). In all the experimental conditions (wild-type Asc1, and S246G and Y333F mutants), the intracellular content of Asc1 substrates as a whole in HeLa cells was not altered after loading with [3H] D-serine (Supplementary Fig. 11h). Efflux via wild-type Asc1 by diffusion was ~19% of that calculated by exchange (Fig. 4). Interestingly, facilitated diffusion in the S246G and Y333F mutants fell to 36%, and exchange to 14% and 23% of the wild-type values, respectively (Fig. 4).
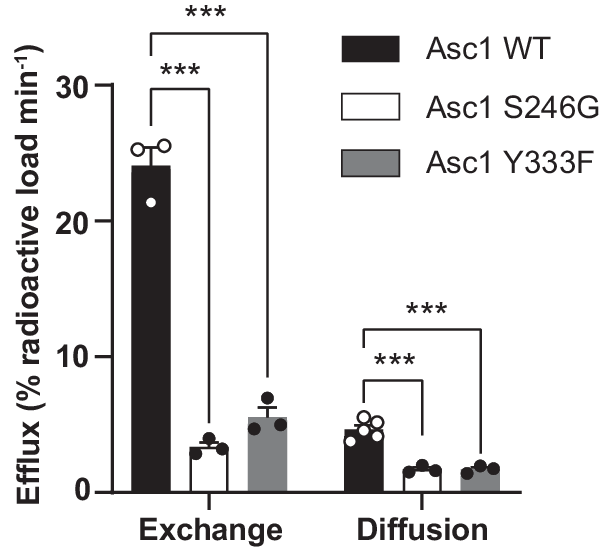
Efflux quantification in percentage of the loaded radioactivity per minute in HeLa cells transfected with wild-type Asc1 or mutants S246G and Y333F. Data (meanâ±âs.e.m.) corresponds to nâ=â5 (WT facilitated diffusion) or nâ=â3 (rest of efflux measurements) independent experiments. A linear model was fitted to transformed data (see material and methods) to obtain p values comparing wild-type Asc1 with each mutant. P values were not adjusted for multiple comparisons. (***) pâ<â0.001. WT vs S246G exchange, pâ=â5.76âââ10â10; WT vs S246G diffusion, pâ=â8.30âââ10â7; WT vs Y333F exchange, pâ=â2.45âââ10â8; WT vs Y333F diffusion, pâ=â7.72âââ10â7.
The first step of the efflux experiments consisted of binding the radiolabeled substrate to the Asc1 transporter in an inward-facing conformation. A conformational transition to outward-facing conformation then took place and substrate was released to the extracellular space. Finally, in the exchange mode, the return to the inward-facing conformation occurred with the external substrate (D-serine) bound to Asc1, whereas this happened in the absence of substrate in the diffusion mode (Supplementary Fig. 11a). By using the mathematical model described in Methods, we estimated the difference between the time the transporter takes to return from the outward- to the inward-facing conformation, either empty (diffusion) or bound to D-serine (exchange) to complete the efflux of 1% of the loaded radioactivity (tIND-tINE), for the wild-type Asc1 and mutants (Table 1, Supplementary Fig. 11g). This time difference was 1.5 and 2.6-fold higher in the S246G and Y333F mutants, respectively, when compared to wild-type Asc1 (Table 1). This observation could be explained by a slower outward to inward transition of the empty transporter (which corresponds to an increase in tIND values) or to a faster outward to inward transition of the substrate-bound transporter (corresponding to a decrease in tINE values), or both. The most likely scenario is a reduction in the velocity of the return of the empty transporter from outward- to inward-facing conformations because mutants S246G and Y333F dramatically decreased efflux by exchange (Fig. 4) and the Vmax of D-serine uptake in HeLa cells (Supplementary Fig. 8f).
In all, our results suggest that Ser 246 and Tyr 333 are relevant both for facilitated diffusion and exchange. PELE analysis predicted the intercalation of substrates between these two residues in the transporter, thus bridging the bundle (TM1 and TM6) and scaffold domains (TM8). Tyr 333 might help coordinate these two domains during the occlusion of the transporter (Fig. 5a). Tyr 333 is unique to Asc1 within the light subunits of HATs, and this observation reinforces the notion that this residue contributes to facilitated diffusion as a mode of transport for HATs of neutral amino acids.
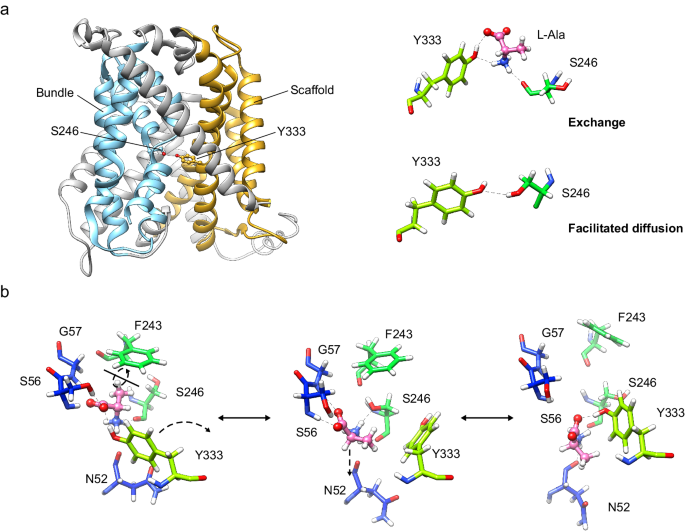
a In the exchange mode of transport, the substrate (L-alanine) intercalates between Ser 246 and Tyr 333, thus bridging the scaffold (tinted in orange) with the bundle domain (tinted in cyan) of the transporter. Analogously, in the facilitated diffusion mode of transport, Ser 246 and Tyr 333 form a potential hydrogen bond between the side chains. Our results support a mechanism in which the connection between Ser 246 and Tyr 333 triggers the fully occluded conformation in the exchange mode of transport, with the transporter bound to substrate, but also in the diffusion mode, where the transporter cycles back with an empty binding cavity. b Proposed role of Ser 246 and Tyr 333 in facilitating the transition of the substrate to and from the transport-productive binding pose. In this model, the productive pose of the substrate between TM1 and TM6 (left, corresponding to the PELE pose 1 shown in Fig. 3b) would be destabilized by a rotamer of Tyr 333 towards TM6 (right, corresponding to the second MD replicate at 24âns), which would have the effect of pulling the substrate away from the GSG motif in TM1, a necessary step for the release of the substrate to the cytosol. The side chain of Ser 246 would stabilize the intermediate step (middle, corresponding to the PELE pose 2 shown in Supplementary Fig. 8a), in which Tyr 333 has rotated but not yet re-bound to the substrate and the substrate itself has experimented aâ~â180° rotation. C atoms in Asc1 residues are colored according with the color of the corresponding TMs as in Fig. 1 whereas substrate C atoms are coloured in pink.
Mechanistic insights into substrate translocation in Asc1
Both our uptake and efflux assays of the Asc1 binding site mutants support the notion that Ser 56, Ser 246, and Tyr 333 are essential for the exchange mode of transport. To further explore the mechanistic role of these residues in amino acid exchange, we performed MD analysis with the L-alanine-bound semi-occluded transporter using the lowest-energy PELE binding pose as the starting point (Fig. 3b).
In two of the three replicates, the substrate remained solidly bound between TM1 (backbones of Ile 53, Ser 56 and Gly 57, as well as the side chain of Ser 56) and TM6 (backbone of Ser 246), whereas Tyr 333 rapidly dissociated from the substrate and showed only a weak interaction (<7% of the frames of the simulation) (Supplementary Fig. 12a). However, in a third replicate, Ser 56 and Tyr 333 competed for the interaction with the substrate α-carboxyl during an initial phase of the simulation (up to ~11âns) (Supplementary Figs. 12b and 13a). This was followed by a rotation of the side chain of Tyr 333 towards TM6 (reminiscent of our PELE pose 2 shown in Supplementary Fig. 8a) that dragged the substrate away from Ser 56 and Gly 57 (Supplementary Figs. 12b and 13b). Strikingly, in the MD simulation, upon the disconnection of the substrate from the unwound region of TM1, the TM1a helix opened further the cytosolic vestibule (Supplementary Fig. 14a, b) and the substrate started to transition out of the binding site (Supplementary Fig. 14c). Once the substrate was definitively dissociated from Ser 56, it remained bound to Tyr 333, as well as to the backbones of Asn 52, Ile 53 (TM1a) and Ser 246 (TM6) (Supplementary Fig. 13b). The interaction with Tyr 333 was lost at 27âns, concomitant with the binding of the substrate α-carboxyl with the ε-amine of Lys 194 (TM5) (Supplementary Fig. 13c). At 30âns, the substrate lost interaction first with Ser 246 and immediately after with Asn 52 and Lys 194 (Supplementary Fig. 13d), at which point L-alanine definitively was released from Asc1.
These results are evocative of previous MD studies of the bacterial LAT BasC33, in which a transition of the substrate towards the equivalent TM5 residue to Asc1 Lys 194 precedes the release of the substrate from the transporter to the cytosol. Functional studies in BasC showed that mutation of this TM5 lysine residue to alanine has a severe impact on the external Km and maximal velocity in BasC and Asc133. Considering the interplay between Ser 56, Ser 246, and Tyr 333 in shuttling the substrate outside the binding site in our MD with human Asc1, we then proceeded to perform a kinetic analysis of [3H] L-alanine uptake to assess whether the mutation of these residues also impacts substrate recognition and translocation (Table 2, Supplementary Fig. 15). Indeed, L-alanine uptake by Asc1 S56A mutant showed an increased extracellular Km and a decreased Vmax compared to wild-type Asc1, thereby suggesting that this mutation impaired the binding energy and the translocation for L-alanine, thus affecting one or several transport-limiting steps. In contrast, the S246G (TM6) and Y333F (TM8) mutants substantially reduced Vmax without affecting Km. This observation thus highlights the role of these two residues in substrate translocation. We further characterized the impact of mutating Ser 246 and Tyr 333, a pair of residues unique in Asc1 among HATs, on the kinetic parameters of transport of D-serine, a substrate uniquely transported by Asc1 among HATs. Much like for L-alanine, S246G and Y333F showed a dramatic reduction in the Vmax of D-serine uptake. Moreover, the S246G mutant also showed a small increment in the Km for D-serine (Supplementary Figs. 8f and 15). This observation suggests that the interaction of the hydroxyl group of Ser 246 with D-serine observed in our PELE analysis contributes to the binding energy of this substrate (Supplementary Fig. 8b).
Taken together, our MD and kinetic analysis point towards a model where Ser 56 would fix the substrate in a transport-productive pose between the unwound segments of TM1 and TM6 (Fig. 5b), thus explaining why the mutation of this residue affects both Km and Vmax, whereas the rotation of Tyr 333 towards TM6 would help to pull the substrate away from Ser 56 in order to transit in and out of this pose. This latter conformation was predicted by the PELE analysis for L-alanine, which also shows that the side chain of Ser 246 interacts with the substrate, perhaps also contributing to the pulling of the substrate towards TM6. Mutations of both Tyr 333 and Ser 246 have a drastic impact on uptake Vmax, thus supporting a mechanistic role of these residues on substrate translocation.