Overview of investigated SAHHs/SIHHs
In this work, SAHHs/SIHHs from archaea, bacteria, and eukaryotes were characterised biochemically (Supplementary Table S2 online). The homologues from M. jannaschii (MjSIHH) and T. maritima (TmSAHH) were previously reported to have SIH cleavage activity14,15,27,35. Structures of archaeal SAHHs were determined from three organisms: Methanococcus maripaludis (MmaSAHH), Pyrococcus furiosus (PfuSAHH)36, and Sulfolobus acidocaldarius (SacSAHH). In addition, the structure of SAHH from Mus musculus (MmSAHH) was determined in complex with inosine.
The substrate range differs depending on the origin of the enzyme
All enzymes were tested in both reaction directions with the hypoxanthine- and adenine-containing substrates. As SAH/SIH cleavage is not preferred in vitro and consequently hard to follow, the reaction was coupled to Hcy S-methylation catalysed by Hcy S-MT (HSMT; EC 2.1.1.10)9 to drive the reaction forward by removing one of the products (Fig. 3b–d). The following analysis with HPLC-DAD allows the direct identification of hypoxanthine- vs. adenine-containing substrate, which is not possible in previously described coupled colorimetric and photometric assays for SAHHs37,38. As a side-product, the corresponding nucleobase (adenine or hypoxanthine) was detected in all set-ups, as previously described39 (Fig. 3b, c). In some reports, SAHH activity was only observed with externally added NAD+40,41,42, while there are also examples showing SAH cleavage and synthesis activity without the addition of extra NAD+32,36,43. All enzymes tested in this work were active without the addition of NAD+.
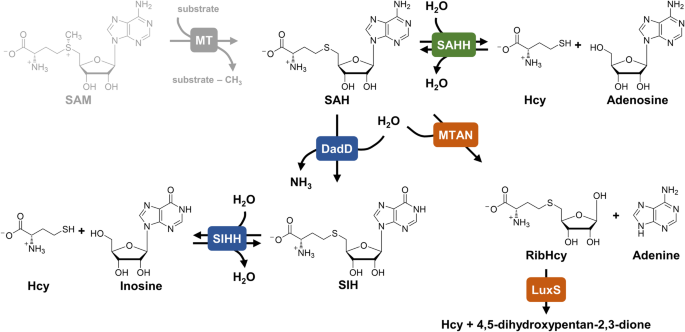
a The sequence-based phylogenetic tree groups the tested SAHHs/SIHHs according to their substrate preference. Nevertheless, this does not fully correspond to the phylogenetic relatedness of the respective organisms: Representatives from Crenarchaeota exclusively accept SAH as substrate; in contrast, most family members tested from Euryarchaeota and a closely related thermophilic bacterium accept both SAH and SIH, including some homologues that clearly prefer SIH. Except for CgSAHH and LlSAHH, bacterial and eukaryotic SAHHs accept both substrates with a substantial preference for SAH. b PfuSAHH-catalysed cleavage of SIH, and (c) SacSAHH-catalysed cleavage of SAH, both with and without addition of ScHSMT (and negative control without enzymes). d The addition of the second enzyme ScHSMT increases the conversion of the cleavage reaction. e The thiol scavenger 5,5’-dithiobis-2-nitrobenzoic acid (DTNB) reacts with Hcy to form a mixed disulphide and 2-nitro-thiobenzoic acid (TNB). For reactions performed at 70 °C it was used instead of ScHSMT to increase the conversion of SAH/SIH cleavage.
Under the chosen conditions, all SAHHs/SIHHs were active for SAH cleavage and synthesis (Table 1), including the homologue from M. jannaschii (MjSIHH) previously described to be specific for SIH as substrate14 (Supplementary Fig. S10; all chromatograms in Figs. S4–S21 online). Nevertheless, this enzyme clearly prefers the hypoxanthine-containing compounds over the adenine-containing ones. The same results were observed for other representatives from Euryarchaeota (MiSAHH, MmaSAHH, PfuSAHH, and TkSAHH). As described before, the bacterial TmSAHH, which is closely related to these euryarchaeal enzymes (Fig. 3a), catalysed SAH and SIH cleavage and synthesis15, in our hands also with a strong preference for the hypoxanthine derivatives. In contrast, another subgroup of SAHHs from euryarchaeal origin (McSAHH, MeSAHH, MhSAHH, and MtSAHH) showed a substantial preference for the synthesis and cleavage of SAH over SIH. In the case of MeSAHH, neither synthesis nor cleavage of SIH was catalysed while the other enzymes of this subset showed slight catalytic activity for SIH synthesis. Comparable results were obtained for the SAHHs from Crenarchaeota (SacSAHH and SsoSAHH), which were only active with SAH or adenosine as substrates. These findings strongly support the assumption that alternative routes for methyl metabolism coupled to purine salvage exist within some classes of Euryarchaeota and closely related bacteria.
In previous work, we successfully used MmSAHH for an in vitro SAM regeneration cycle with alternative nucleobases including hypoxanthine44. This activity was now confirmed in the individual cleavage and synthesis reactions; however, reactions with SAH were in this case preferred. A similar pattern was observed for the SAHH from the bacterium Pseudomonas aeruginosa, while neither the representative from Corynebacterium glutamicum nor from the plant Lupinus luteus accepted inosine or SIH as substrates. As SIH had been described as a metabolite in S. flocculus, along with the presence of an SAH deaminase16,17, we tested SAHHs from two Streptomyces species (SaSAHH and SfSAHH). In both cases a low activity for SIH synthesis was detected; however, no activity for SIH cleavage (Supplementary Figs. S17 and S18 online). It might be that SIH is metabolised by other enzymes in these bacteria, suggesting an alternative SAH metabolism pathway than the one described for Euryarchaeota and archaeal-type bacteria.
Increased temperatures do not influence the substrate preferences
So far, the substrate preferences of archaeal, bacterial, and eukaryotic SAHH/SIHH representatives were tested at 37 °C to investigate their applicability in a multi-enzyme SAM regeneration cycle44. Some of the archaeal (PfuSAHH, TkSAHH, McSAHH, MiSAHH, MtSAHH, MjSIHH, SacSAHH, and SsoSAHH) as well as archaeal-type (TmSAHH) enzymes are derived from (hyper-)thermophilic organisms. TmSAHH produced and purified at room temperature was previously described to require thermal activation to attain enzymatic activity27,42. However, in our hands TmSAHH, as well as the other thermophilic enzymes, was enzymatically active without previous thermal activation. Assays to test the influence of higher temperatures on the substrate preferences were performed at 70 °C in SAH/SIH synthesis and cleavage directions with two selected model enzymes, PfuSAHH (Euryarcheota) and SacSAHH (Crenarchaeota). For reactions performed at 70 °C, HSMT from Saccharomyces cerevisiae (ScHSMT) was replaced by 5,5’-dithiobis-2-nitrobenzoic acid (DTNB). It acts as thiol scavenger reacting with Hcy in stoichiometric amounts to form a mixed disulphide and 2-nitro-thiobenzoic acid (TNB), and therefore shifts the reaction equilibrium to the product side37,45 (Fig. 3e). The suitability of DTNB as substitute for ScHSMT was tested beforehand at 37 °C (Supplementary Fig. S22 online). At 70 °C, PfuSAHH still showed a clear preference for SIH synthesis and cleavage compared to SAH (Supplementary Fig. S23 online). For SacSAHH, the conversion with the unfavoured substrates was slightly higher compared to the reactions performed at 37 °C; nevertheless, the enzyme still showed a substantial preference for SAH synthesis and cleavage (Supplementary Fig. S24 online). This strongly indicates that the tested SAHHs/SIHHs show the same substrate preferences at both temperatures. Increased conversion of SacSAHH-catalysed SIH cleavage suggests that they could degrade small amounts of SIH under higher living temperatures.
Different motifs for binding the nucleobase of the substrate/product in the subgroups of archaea
The differences in substrate range among the investigated SAHHs/SIHHs prompted us to conduct a detailed sequential and structural comparison to identify the underlying molecular basis. A sequence alignment of the enzymes tested in this work (Supplementary Fig. S26 online), showed that MmSAHH as well as PaSAHH are missing a 40 amino acid segment in the catalytic domain as opposed to the other eukaryotic and bacterial representatives. All investigated archaeal SAHHs/SIHHs, except for MeSAHH and MhSAHH, also lack this structure segment as described before12,33. Another sequence feature discriminating MeSAHH and MhSAHH from the other archaeal and archaeal-type homologues is the length of their C-terminus. The majority of SAHHs/SIHHs analysed in this work containing a shortened C-terminus are (hyper-)thermophilic, confirming the assumption36 that the length of the C-terminus correlates with the thermophilicity of the enzymes46. Based on our sequence analyses, the fingerprint motif suggested to distinguish extremophile SAHHs from mesophilic enzymes19 can be further specified: Crenarchaeota and a large part of Euryarchaeota feature HxTxE as a signature, this matches the sequence signature of bacterial and eukaryotic representatives [HxTxE(Q)]. A subgroup of the euryarchaeal and archaeal-type bacterial SAHHs, including six homologues analysed in this study have an HxExK motif (Fig. 4). Based on the data on euryarchaeal sequences available in the UniProt database, this subgroup encompasses mainly the Methanococci, Thermococci, Methanobacteria, Hadesarchaea, Archaeoglobi and Methanosarcina; while euryarchaeal enzymes from various other classes (e.g. Methanocella and Methanophagales) feature the HxTxE motif as found in Crenarchaeota; in further classes (e.g. Methanomicrobia and Theionarchaea), there is no clear trend visible. Our experiments show that the homologues with a HxExK motif prefer SIH as substrate while enzymes with a HxTxE(Q) motif prefer SAH. These results infer that the fingerprint motif distinguishes SAH-preferring SAHHs from SIH-preferring SIHHs.

Partial alignment of the amino acid sequences of SAHHs/SIHHs tested in this study. The fingerprint motifs are highlighted in grey while the other residues are coloured by conservation according to the BLOSUM62 algorithm.
Genomes from Euryarchaeota and archaeal-type bacteria encode a deaminase
In order to get more insight in the potential alternative SAH salvage pathway, a BLASTP47 search for a protein homologue of DadD from M. jannaschii (MjDadD, Fig. 1) in the organisms producing the SAHHs/SIHHs analysed in this work was performed. The genomes from all analysed Euryarchaeota, as well as from the archaeal-type bacterium T. maritima, were found to have a homologue encoded (Supplementary Table S6 online). This is inconsistent with the substrate scope of characterised archaeal SAHHs/SIHHs as the subgroup of the tested euryarchaeal McSAHH, MeSAHH, MhSAHH and MtSAHH showed no activity for SIH cleavage. Notably, the BLASTP search revealed multiple isoforms of MeSAHH (UniProt accessions: D7E8L4 and D7E8N1) and MhSAHH (A0A1L3PZV2) suggesting that those organisms contain more than one SAHH, with putatively different substrate preferences, as inferred from their fingerprint motif. For McSAHH and MtSAHH, no isoforms were found. In these organisms, SIH resulting from DadD catalysed SAH deamination needs to be degraded via a pathway without SAHHs/SIHHs. Hence, a BLASTP search for homologues of Escherichia coli MTAN (EcMTAN), cleaving SAH into adenine and S-ribosyl-l-homocysteine, was performed. Only C. glutamicum, T. maritima and L. luteus were found to encode an MTAN homologue (Supplementary Table S7 online). This suggests that in the organisms encoding a DadD and an SAHH only cleaving SAH, SIH is metabolised by another pathway. Moreover, the lack of MTAN homologues in Euryarchaeota encoding an SAHH preferring SIH but also accepting SAH as substrate, strongly indicates that those enzymes have a dual function for SIH and SAH metabolism. The organisms without an encoded SAH deaminase do not need their SAHH to degrade SIH in addition to SAH. A more detailed BLASTP search including all available entries in the UniProt database48 resulted in the identification of further homologues of MjDadD, exclusively within the domain of bacteria and the phylum of Euryarchaeota. This suggests that the alternative SAM regeneration pathway via SIH is indeed not present in all archaea, but only in a subgroup from the phylum of Euryarchaeota and closely related thermophilic bacteria. Due to their shared living environment, thermophilic bacteria were shown to have obtained genes via horizontal transfer from archaea49. The reason for substantial SIH degradation and synthesis by MmSAHH and PaSAHH remains yet unclear. Likely, there will be other amino acid residues involved in substrate recognition which are not obvious from the sequence alignments.
Architecture and domains of archaeal SAHHs
Thus far, the sequence comparison revealed differences concerning the constitution of the catalytic domain, the length of the C-terminus, and the composition of the signature motif binding the nucleobase moiety of substrates and products. A detailed structural analysis was performed to determine how those differences influence the three-dimensional structure of SAHHs/ SIHHs and effect the substrate preferences. The archaeal enzymes were an ideal model system as they show different substrate preferences; however, no structures of archaeal SAHHs have been published to date. Here, we present the structures of three archaeal SAHHs (PfuSAHH, MmaSAHH, SacSAHH); in addition, the structure of the eukaryotic MmSAHH was determined in complex with its unusual substrate inosine (Table 2). In all SAHH structures obtained, the NAD+ cofactor was present indicating that it was co-purified with the enzyme.
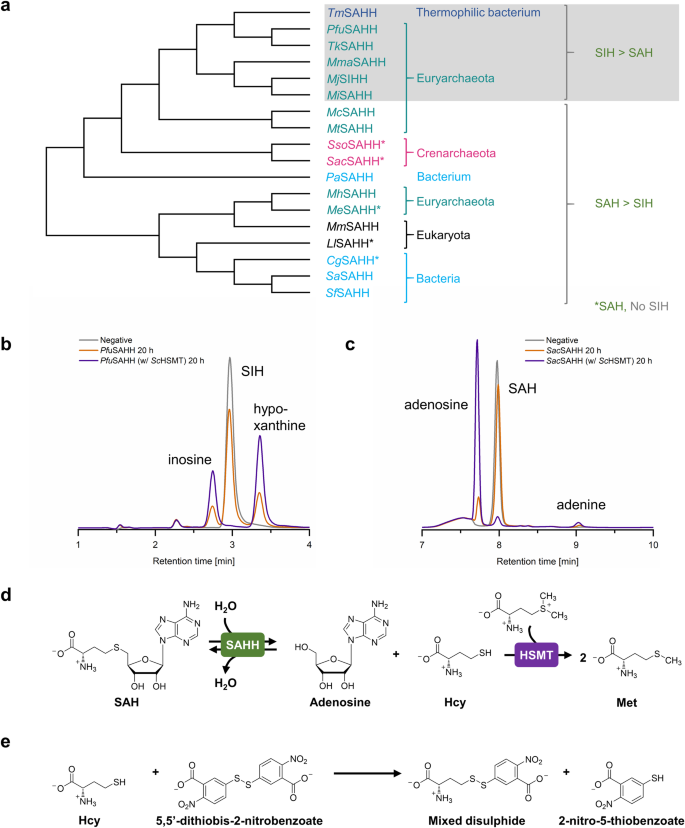
The archaeal SAHH monomer contains the same three domains as previously studied SAHHs across other domains of life: substrate-binding domain, cofactor-binding domain, and C-terminal domain27,32,34,42,50. The binding modes of NAD+ at the cofactor-binding domain and inosine or adenosine in the substrate-binding domain of archaeal SAHHs are similar to those observed in other SAHHs of distinct origin (Fig. 5a–d)26,28,31,32,34,51,52. In this study, the nucleoside moieties of the three ligands, adenosine, inosine, and SIH, participate in a similar pattern of hydrogen-bonding and non-bonding interactions (Supplementary Table S5 online). Evaluating the euryarchaeal structures elucidated in this study, it supports the assumption that the Glu57 residue (in the HxExK motif, numbering according to PfuSAHH, Fig. 5c/d and S27A/B online) forms a hydrogen bond with the heterocyclic nitrogen atom N1 of the nucleoside, analogous to the Thr residue in crenarchaeal, bacterial and eukaryotic homologues (in the HxTxE motif, e.g. Thr53 in SacSAHH, Fig. 5a and S27D online). The Lys residue at the end of the motif is found in enzymes that prefer hypoxanthine-containing substrates; here, the positively charged side chain can form a hydrogen bond with the oxygen at C6 in inosine, as seen in the structures of PfuSAHH (Lys59 in Fig. 5e and S27A/B online) and MmaSAHH (Lys74 in Supplementary Fig. S27C online), contributing to the stabilisation of the substrate in the active site. The motif HxTxE found in SacSAHH (Crenarchaeota) shows the same interactions with the substrate adenosine as in bacterial and eukaryotic enzymes19. As described before19, we observed that the exo-amino group of adenosine bound in SacSAHH forms additional hydrogen bonds with the carbonyl oxygen atoms in the main chains of Glu342 and His344 indicating that the adenine ring is in its preferred tautomeric amino form. Regarding the structures of PfuSAHH and MmaSAHH with their preferred substrates inosine or SIH, the oxygen attached to C6 seems to form additional hydrogen bonds with the carbonyl oxygen atoms of Asp348 (PfuSAHH) or Asp362 (MmaSAHH) in the main chains. This suggests that the hypoxanthine ring is in the imino-hydroxy form (Fig. 5f), although inosine has been shown to be most stable in the amino-oxo form in water53. Analysis of superposed structures of MmSAHH with bound adenosine54 and bound inosine showed no substantial differences in the architecture of the substrate binding pocket depending on the bound substrate (Supplementary Fig. S28 online).
a Mode of adenosine and NAD+ cofactor binding in the active site of SacSAHH (PDB ID: 7R39). b Mode of inosine and NAD+ cofactor binding in the active site of MmaSAHH (PDB ID: 7R3A). c Mode of SIH and NAD+ cofactor binding in the active site of PfuSAHH (PDB ID: 7R38). d Mode of inosine and NAD+ cofactor binding in the active site of PfuSAHH (PDB ID: 7R37). e Inosine is bound by the motif HxExK in the substrate-binding domain (cartoon representation; PDB ID: 7R37). f Tautomers of inosine. Ligands are represented as sticks.
Overall conformational states and the molecular gate of archaeal SAHHs
Different conformational states of SAHHs have been described, where the open conformation was observed with no bound substrate/product in the substrate-binding domain, while the protein with a bound substrate or analogue shows the overall closed conformation. An example is a structure (PDB ID: 4LVC) of a bacterial SAHH showing adenosine bound in three subunits displaying a closed conformation while the fourth, ligand-free, subunit is in an open conformation28. All archaeal SAHH complexes in this study also display a closed conformational state including the PfuSAHH complex with SIH bound as substrate.
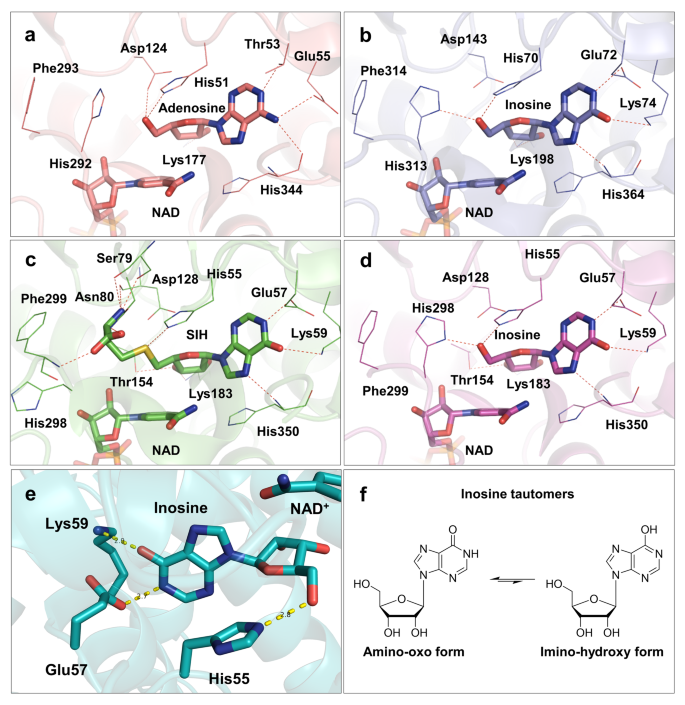
In addition to the closed overall conformation of the PfuSAHH•NAD•SIH complex, the molecular gate loop displays a flexibility. The transitions between the overall conformation and the conformation of the gatekeeper residues are independent from each other. Yet, His298 and Phe299 forming the molecular gate in PfuSAHH can only act as gatekeepers in the closed conformation when substrate-binding domain and cofactor-binding domain form the channel, as described before28,31. Interestingly, the His298 residue alone displays a noticeable conformational difference in this study. In the inosine bound complex, His298 is situated within the active site staging a His-IN conformation (gate shut) where its imidazole ring is oriented towards the γ-carboxylate group of Asp128 with a distance of 4.2 Å. The Nδ1 group of His298 is oriented to the O5´ group of inosine with a distance of 2.7 Å. A similar pattern is observed for MmaSAHH and SacSAHH complexes. However, the rotamer conformation of His in the His-IN state of SacSAHH deviates from the rest of the His-IN conformations among the archaeal SAHHs (Fig. 6) in this study. This indicates that there are not only different His orientations possible in the His-OUT state55, but also in the His-IN state. Comparison of the MmSAHH structures with bound adenosine and inosine (Supplementary Fig. S29 online) and the structure of SacSAHH suggests that the rotamer conformation is not dependent on the bound nucleoside substrate. The His298 residue of PfuSAHH•NAD•SIH complex swings away from Asp128 by 10.4 Å and forms a hydrogen bond with the carboxylate group of Glu302 with a distance of 2.7 Å signifying a His-OUT conformation leaving an open molecular gate. In a previously described structure of the Lupinus luteus SAHH32, the channel is open (His-OUT) even though adenosine is bound, while in the SAHH structure from Mycobacterium tuberculosis the same trend is seen as for PfuSAHH with the channel closed (His-IN) with adenosine bound and channel opened with SAH bound31. Similarly, our complexes of PfuSAHH and MmaSAHH with inosine and the complex of SacSAHH with adenosine show a closed conformation state of the molecular gate in their active site (Fig. 6). In accordance with Manszewski et al., comparison of our structures with already published SAHH structures did not lead to an apparent correlation between the conformation of the gate residues and the bound substrates56. Instead, the state of the reaction catalysed by SAHHs may determine the conformation of the gatekeeper residues to protect the unstable 3´-keto intermediates from exposure to the aqueous environment, as proposed by Yang et al57.

The His residue states are represented as His-IN (IN) and His-OUT (OUT). a In the PfuSAHH•NAD•inosine complex (PDB ID: 7R37), the His gatekeeper residue is in the IN orientation thereby closing the channel entrance. b In the PfuSAHH•NAD•SIH complex (PDB ID: 7R38), the gatekeeper residue forms an OUT orientation leaving the channel entrance open. c For the MmaSAHH•NAD•inosine complex (PDB ID: 7R3A) the gatekeeper residue is in the IN orientation. d In the SacSAHH•NAD•adenosine complex (PDB ID: 7R39), the gatekeeper residue is in the IN orientation that makes the channel entrance shut and inaccessible. The side chain of the gatekeeper residue of SacSAHH (circled red) shows another rotamer conformation than in PfuSAHH and MmaSAHH.
Differences in archaeal compared to bacterial and eukaryotic SAHHs
While the protein sequences of archaeal SAHHs/SIHHs are highly conserved (Supplementary Fig. S32 online), the substrate range differs within the distinct phyla of archaea. The PfuSAHH•NAD•inosine and MmSAHH•NAD•inosine complexes determined in this work were used for structural comparison with the bacterial PaSAHH (PDB ID: 6F3N). Besides the different signature motifs binding the nucleobase moiety, the overall architecture of the macromolecular environment of active sites is comparable between those three enzymes (Fig. 7a). The PfuSAHH•NAD•inosine complex lacks a monovalent cation (Na+ for MmSAHH and K+ for PaSAHH) coordinated by the hinge element in proximity to the nucleobase binding pocket34,52 (Fig. 7b–d). The absence of a comparable monovalent cation was observed in all structures of archaeal enzymes. Therefore, the catalytic activity of archaeal enzymes might be independent from the presence of monovalent cations as previously seen for a cyanobacterial homologue from Synechocystis sp. PCC 680352.
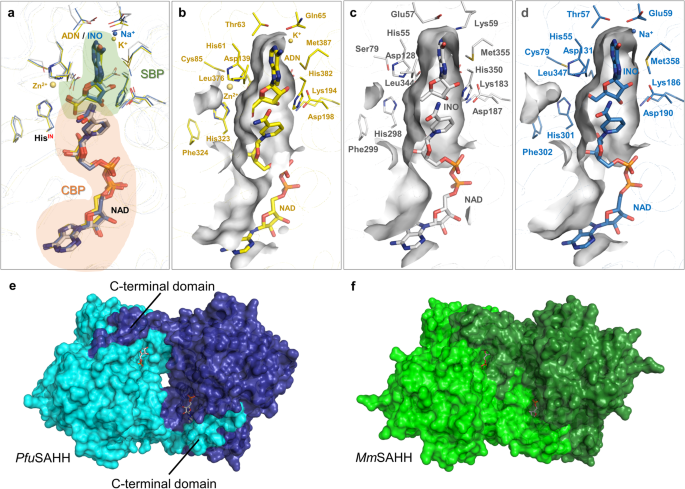
a Superposition of the active sites of SAHHs from bacterial (yellow), eukaryotic (blue), and archaeal (white) organisms with bound substrates and NAD+ cofactors (SBP: substrate-binding pocket; CBP: cofactor-binding pocket). b SBP of bacterial PaSAHH in complex with adenosine (ADN). Zn2+ and K+ are shown as yellow spheres (PDB ID: 6F3N34). c SBP of archaeal PfuSAHH in complex with SIH (PDB ID: 7R38). d SBP of eukaryotic MmSAHH in complex with inosine (INO). Na+ is shown as blue sphere (PDB ID: 8COD). e In PfuSAHH, the cofactors are loosely covered by the adjacent monomer while some room is visible between the monomers, indicating a less stable enzyme (PDB ID: 7R38). f In MmSAHH, the cofactors are tightly covered by the C-terminus of the adjacent monomer (PDB ID: 5AXA53).
Looking at the overall structure of archaeal SAHHs compared to their eukaryotic and bacterial counterparts, the major difference is the shortened C-terminus, which interacts with the cofactor bound to the neighbouring subunit. Due to the shortened C-terminus, the archaeal enzyme structures lack hydrogen bonds, which are formed between amino acids (Lys426 and Tyr430) of one subunit with the 2´- and 3´-hydroxy groups of the adenosine moiety and the pyrophosphate of NAD+ bound in the adjacent subunit, present in MmSAHH and other mesophilic homologues. Instead of interactions between C-terminal domains of neighbouring monomers, the major forces for stabilisation are provided by aromatic and hydrophobic amino acid residues at the interfaces of the tetramers36. In addition, the interfaces between the monomers of the archaeal SAHHs show a cleft, when looking at the surface representation (Fig. 7e). This is not visible in eukaryotic SAHHs, such as the one from mouse (Fig. 7f); also because of the longer C-terminus that covers the cofactor. This means the cofactor is more exposed to the environment in archaea, and indicates a less stable tetrameric form for archaeal SAHHs/SIHHs, as fewer interactions are found between the monomers. TmSAHH was found to require a much higher concentration of NAD+, which was linked back to the shortened C-terminus, impacting the binding affinity27. TmSAHH has the shortest C-terminus, which is three amino acids shorter than the archaeal ones. All SAHHs/SIHHs in this work (including TmSAHH) were active without the addition of NAD+ at 37 °C and thus no dependency between concentration of NAD+ and length of C-terminus was observed in our hands. Moreover, recombinant TmSAHH was described to only be enzymatically active after a heat-induced conformational change27,42. To further investigate this observation, we used recombinant enzymes either previously preincubated at 95 °C or permanently handled at room temperature for crystallisation of the hyperthermophilic PfuSAHH in complex with inosine. Comparison of both structures showed no substantial differences in the conformation of the differently treated enzymes (Supplementary Fig. S30 online). Independent of the previous thermal treatment, PfuSAHH, as the other archaeal homologues, formed stable homotetramers in the crystal. To verify the arrangement of individual units in the multimeric assembly of the archaeal SAHH crystal structures, tetramers were constructed and analysed for each SAHH individually in their respective space groups using PDBePISA58. The prediction analysis of PfuSAHH tetramers (both inosine and SIH complexes) showed an average value for the solvation free energy on formation of the assembly (ΔGint) of −129.0 kcal mol−1. Similarly, MmaSAHH tetramers and SacSAHH tetramers exhibited an average ΔGint value of −133.5 kcal mol−1 and −127.5 kcal mol−1, respectively. In comparison, selected eukaryotic SAHHs with a long C-terminus, such as MmSAHH tetramers (both inosine and adenosine complexes), showed an average ΔGint of −186.7 kcal mol−1. These predictions suggest that the eukaryotic SAHH tetramers display more favourable solvation free energy than the archaeal SAHH tetramers. Regarding the stability of archaeal SAHH tetramers, the shortened C-terminal domain was speculated to either be useful for intramolecular motion at high temperature and prevent denaturation based on a protein model for PfuSAHH36, or to positively affect the thermostability of the enzyme27. This is in accordance with the analyses provided in this study.