Materials
β-Cyclodextrin (Merck, Germany), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 2,2-Diphenyl-1-picrylhydrazylradical (DPPH), 2,2′-azinobis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), and dimethylsulfoxide (DMSO) were purchased from Merck company. The Silibinin and Zein were provided by Golexir (Mashhad, Iran) and Sigma Aldrich (France) companies, respectively. The human colon (HT-29), gastric (AGS), and normal umbilical vein endothelial cells (HUVEC) were provided by the Pastore Institute of Iran.
SZBC-NCs synthesis
To produce the silibinin-loaded Zein-β cyclodextrin nano-carriers (SZBC-NCs), the nanocarriers’ ingredients were separately dissolved in an appropriate solvent. In this regard, Zein and β-cyclodextrin were dissolved in ethanol (40% V/V) and stirred for 1 h at 50 °C. Then, Silibinin (20 mg) was added to the ethanolic β-cyclodextrin solution. Finally, the cooled solution was dropwise added to the Zein solution under a probe-mediated ultrasound homogenizer device at 300 Watt-power for a 10-min sonication process (8ʹʹ On 2ʹʹ Off). The resulting solution was centrifuged, rinsed with distilled water, and lyophilized applying a − 80 °C—Freez drier (Scheme 1).
Producing the SZBC-NCs process.
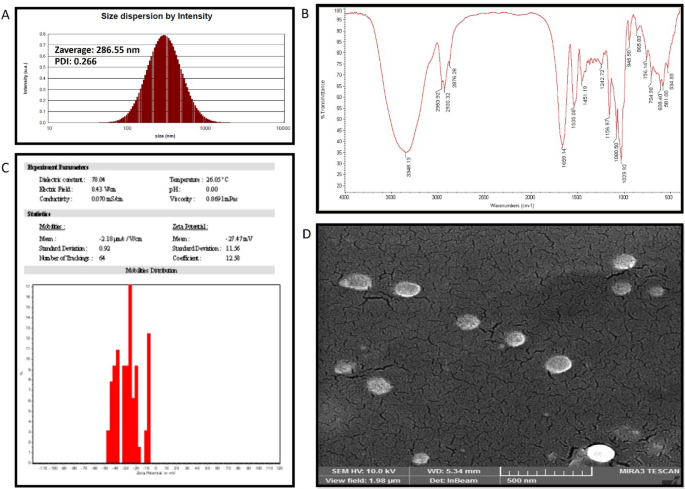
SZBC-NC characterization
The SZBC-NCs’ hydrodynamic and dehydrated size dimensions were performed by dynamic light scattering (DLS) (Zetasizer (nanoparticle SZ-100)) and Field emission scanning electron microscopy (FESEM) techniques, respectively. The DLS analysis was conducted in physiologic pH at 37 °C conditions. Also, the FESEM study was performed by drying a 50-µL sample drop on a clean piece of softened aluminum foil and coating it with a thin ionic gold layer before electron microscopy imaging. The SZBC-NCs chemical composition was characterized by conducting FTIR analysis. To this purpose, 200 mg of potassium bromide (KBr) was added to SZBC-NCs (2 mg) and compressed to a thin disc for scanning the FTIR spectrums in the 4000 to 400 cm−1 wavenumbers interval at 4 cm−1 resolution. Finally, the nanocarriers’ surface charge was evaluated by applying a Zetasizer device (nanoparticle SZ-100).
Silibinin loading and releasing efficiency
The Silibinin loading efficiency into the Zein-beta cyclodextrin nanocarriers was determined utilizing a visible-ultraviolet spectrophotometer (Hutch, USA). Briefly, the standard curve of the silibinin was provided recording the absorbance of consent concentrations at 225 nm. The silibinin loading efficiency was estimated by recording the silibinin absorbance before and after the loading process and calculating its concentration considering its provided standard curve. Also, the drug release rate was measured by mixing SZBC-NCs (100 mg) in a phosphate buffer solution and placing it in a pre-activated dialysis bag, which was suspended in a beaker of phosphate buffer. Then, the dialyzed silibinin absorbance was recorded at 225 nm after 6, 12, 24, 48, 72, 96, 120, and 144 h of incubation at 37 °C. Finally, the Silibinin release graph was plotted.
SZBC-NC antioxidant activity: ABTS and DPPH assay
To measure the SZBC-NCs’ antioxidant activity, their radical scavenging ability was measured as previously described20. Briefly, the activated ABTS and DPPH solutions were prepared. The ABTS solution (7 mM) was activated by potassium persulfate (2.45 mM) solution at 1:1 proportion, diluted with water (1:1 V/V), and stored for 14 h at 25 °C in dark conditions. Also, the ethanolic DPPH solution was prepared by dissolving 1 mg of DPPH in 17 mL of ethanol. The antioxidant assay was conducted by separately adding 5 µL of different concentrations of SZBC-NCs mixture (62.5, 120, 250, 500, and 1000 µg/mL) to 3.995 mL of both ABTS and DPPH solutions. The final solutions were kept at room temperature for 30 min in dark conditions. The ABTS and DPPH inhibition rates were measured by recording their related absorbance and the samples absorbance was recorded at 734 and 517 nm, respectively20. The inhibition rate of ABTS (IRA%) and DPPH (IRD%) was calculated as the following equation:
$$\text{IRA\% or IRD\%}= \left(\frac{\left[A\right]c-[A]s}{\left[A\right]c}\right)\times 100.$$
MTT assay
Three different cell lines (HT-29, AGS and Huvec) were purchased from the cell bank of Ferdowsi University of Mashhad (Iran). The cancerous (HT-29 and AGS) and normal Huvec cell lines were seeded (5 \(\times \) 103 cells/cm2) in a complete DMEM cell culture medium and cultured for 24 h at the standard conditions (5% CO2, 37 °C, and 95%humidity). The medium was supplemented with FBS (10%) and streptomycin/penicillin (100 mg/mL/100 U/mL). The 24-h cultured cells were seeded (at 5 \(\times \) 103 cells/well density) and incubated for 24 h in 96-well plates. Then, the cells were treated with different SZBC-NCs concentrations (7.8, 15.6, 31.2, 62.5, 125, 250, and 500 µg/mL) for 48 h. Then, the MTT (0.5 mg/mL)-containing medium was replaced and incubated for 3 h at 37 °C. To dissolve the produced formazan, DMSO was added to the wells to record the formazan absorbance at 570 nm (Stat fax 2100 plate reader). The cells’ survival was calculated considering the following equation:
$$\text{Cell survival }(\text{\%}) = \left(\frac{\left[A\right]s}{\left[A\right]c}\right)\times 100.$$
DAPI staining
The 48-h exposed cells with different doses of SZBC-NPs (205, 225, and 255 µg/mL) were fixed with paraformaldehyde (4%) for 9 min. The PBS and triton X-100 (0.1%) solutions were used for the rinsing and permeabilizing process of the cells. Finally, the fixed-permeabilized cells were stained with DAPI (4,6-diamidino-2-phenylindole) for 5 min. The cells were studied by a fluorescent microscope (Olympus IX81 invert fluorescence microscope), which was equipped with an Olympus DP70 camera (Olympus Corp., Tokyo, Japan)21.
Gene expression profile
The 48-h exposed HT-29 cancer cells with different doses of SZBC-NCs (205, 225, and 255 µg/mL) were harvested for the RNA extraction process. The cells’ total RNA was extracted by utilizing an RNA extraction kit (Pars Tous, Iran) and prepared for synthesizing the cDNA libraries applying a cDNA synthesis kit (Pars Tous, Iran). The Caspase 3, Caspase 9, and Nf-KB gene primer sets were designed by Allel ID6 software applying the exon junction method (Table 1). The target genes’ cDNA was amplified by PCR technique (Bio-Rad CFX96). Also, the Q-PCR method was applied to measure the gene expression profiles compared with the control house-keeping gene (GAPDH) applying the 2−∆∆CT method for calculating the target genes’ fold change values. A SYBR green-supplemented PCR master mix (Qiagen, Hilden, Germany) was used for conducting Q-PCR analysis. Finally, a comparative threshold cycle method was conducted to normalize the fold change values.
Statistical analysis
The statistical measurements were conducted by applying SPSS-20 software. The One-way ANOVA statistical analysis was utilized to determine the statistical significance levels of p-values. The less than 0.001, 0.01, and 0.05 were considered as the statistically significance levels expressed as ***, **, and * index.