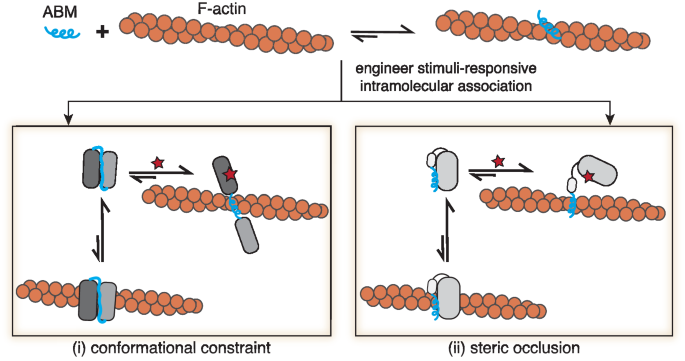
Engineering CASTs to control F-actin binding
Native ABM sequences spontaneously adopt active conformations that engage with F-actin (Fig. 1). Recently solved Cryo-EM structures for the ABM, Lifeact, show an active α-helical structure bound to F-actin at an n/n + 2 site near F-actin’s D-loop18,40. Active ABM conformations, such as Lifeact’s, represent more energetically stable structures in the presence of F-actin compared to inactive states. To construct F-actin protein switches, we sought to re-engineer ABM structures, such that they adopt inactive, yet stable, states. We reasoned that short ABMs would be more sensitive to structural changes compared to larger ABDs since ABMs lack buffering amino acids or domain architectures, offering an opportunity for rational-based design. To inactivate ABMs, we settled on an approach where intramolecular binders are placed on the N- and C-termini of the ABM sequence. Termini binding would lead to a stable state that conformationally constrains or occludes the ABM, rendering it inactive (Fig. 1). Strong intramolecular binding can then be disrupted by stimuli, such as peptides, small molecules, or light, that relieve the constraint on the ABM and favor the active form of the ABM structure. Here, we focus on engineering two native ABMs, ZO-1’s ABS and Lifeact (Supplementary Fig. 1a), the latter a higher affinity ABM sequence toward F-actin and a widely used F-actin marker in cells35, into CASTs. With our strategy, the ABM’s active state would be perturbed in the switch’s “off” state, and upon activation, a conformational change would ensue, leading to an unconstrained conformation and resulting in the ‘on’ state. To directly translate our approach to living cells, we screened CAST candidates in cells using microscopy and co-stained for F-actin to characterize the CAST’s properties.
Peptide-based CASTs for long timescale activation
For our first CAST design, we took advantage of heterospecific synthetic peptides, SynZips (SZs), that form coiled-coil interactions with their peptide partners41. SZ4 binds to either SZ3 or SZ21, albeit with a significantly higher affinity for SZ21. We envisioned a design, where SZ3 and SZ4 bind intramolecularly (Fig. 2a), which would disrupt the active form of the ABM constrained between them. Expression of SZ21, the stimulus in this case, would outcompete intramolecular binding, relieving the constraint on the ABM and leading to the F-actin association. Modeling predicted a parallel coiled-coil for the interaction between SZ3 and SZ4 and a C- to N-terminal distance of 7.4 nm (Fig. 2b). This distance must be spanned by the ABM and any adjacent amino acids to allow for intramolecular binding. Since ABMs are short peptide sequences, we considered three different scenarios (Fig. 2c): (i) the ABM length alone does not span the 7.4 nm distance, leaving the ABM in the active state, (ii) a linker sequence on either end of the ABM accommodates the 7.4 nm distance, inactivating the ABM by constraining its conformation, and (iii) additional linker sequences span the required distance for intramolecular binding but do not result in ABM inactivation. To estimate the amino acid lengths necessary to span the C- to N-terminal distance, we applied a worm-like chain (WLC) model of linker sequences to predict the end-to-end distance distributions42,43 for both ZO-1’s ABS and Lifeact (Fig. 2d). Probabilities over 0.01 were observed for linker lengths with >50 amino acids (AAs). Accordingly, we designed and cloned long-linker SZ-based CAST constructs containing 0–9 five AA-long linkers on both sides of the ABMs, pep0-pep9, for ABS and Lifeact (Fig. 2d).
a Schematic of a peptide-based CAST system. The ABM is initially constrained (gray) in an “off” state by two SynZip sequences, SZ3 and SZ4, which form a coiled-coil interaction intramolecularly. After the introduction of a peptide stimulus (SZ21), which outcompetes SZ3 for SZ4 binding, a transition to the active “on” state (blue) capable of F-actin binding occurs. b AlphaFold2 model prediction of SZ3:SZ4 complex with a C-to-N-terminal distance of 7.4 nm. c Structural considerations for the initial “off” state of CASTs. The C-to-N-terminal distance of the SZ3:SZ4 complex must be spanned by the interdomain residues. Flexible linkers can be installed on either end of the ABM to increase interdomain length. Short linker lengths impede SZ3:SZ4 binding, allowing the ABM to remain active (left). Long-linker lengths, on the other hand, lead to SZ3:SZ4 binding but no constraint on ABM (right). Only the optimal linker length will allow for both SZ3:SZ4 interaction and a conformational constraint on the ABM (middle), resulting in the “off” state. d Probability distributions of end-to-end terminal distances for ABS and Lifeact pep0-pep9 (see bottom) with varying interdomain residue lengths from a worm-like chain (WLC) model (top). The primary sequence of peptide-based CAST candidates for control of F-actin binding (bottom). Source data are provided as a Source Data file.
To examine ABM activity, we expressed pep0-pep9 designs in HeLa cells, which present abundant actin filaments within stress fibers and lamellipodial structures44,45, and stained for endogenous F-actin using phalloidin (Fig. 3a, c and Supplementary Figs. 1b, 2). We observed similar F-actin engagement for short and long-linker pep designs compared to ABM-only controls, as was anticipated from the structural demands of the SZ3-SZ4 interaction (see above). However, for intermediate linker lengths, we observed robust inactivation of ABM F-actin-binding activity. Using the phalloidin stain as a mask, we compared the fluorescence intensities of the pep0-9 designs on F-actin to the intensities in the cytoplasm and calculated a percent inhibition of F-actin engagement compared to the native ABM’s localization (Fig. 3b, d). Designs with >50% inhibition were considered inactive. For both ABS and Lifeact, the pep6 designs, containing 60 AA linker residues, yielded the highest levels of inactivation and were selected for further development (termed pepCAST).
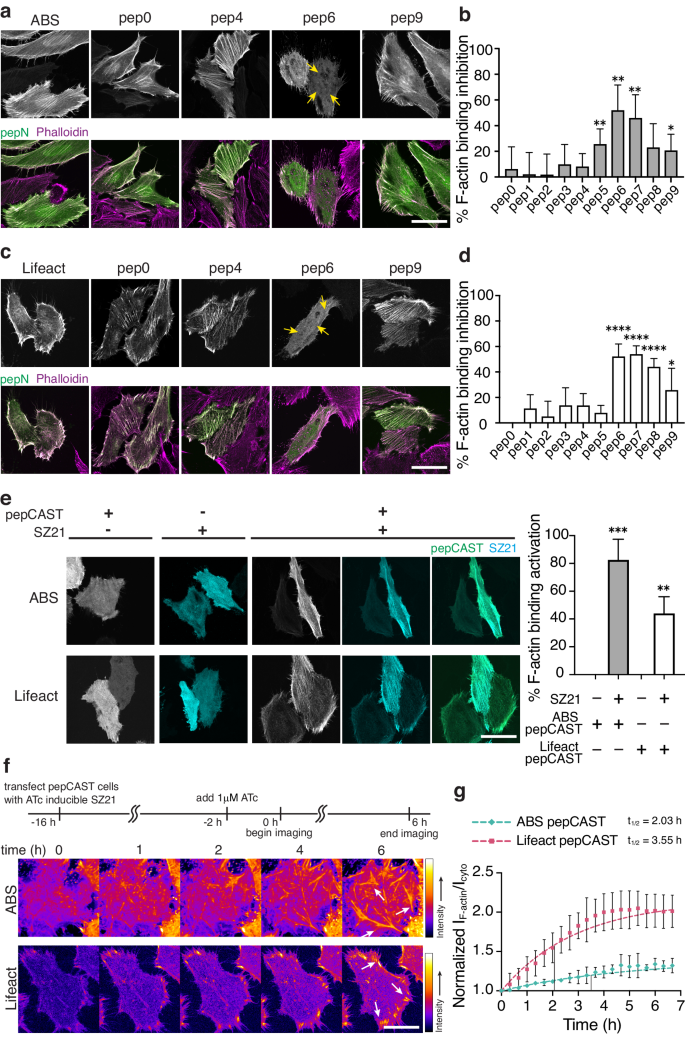
a Fluorescent micrographs of fixed HeLa cells expressing ABS pep0-pep9 in the absence of SZ21. Total cellular actin was visualized by Phalloidin staining (magenta). ABS pepCAST (pep6) was selected for further study since its localization to F-actin was significantly reduced (yellow arrows). Scale bar, 30 µm. b Quantification of F-actin binding inhibition for ABS pep0-pep9, where 0% is defined by ABS-only localization and 100% is defined by GFP-only localization. Bars represent mean ± SD. n = 40 biological replicates. c Fluorescent micrographs of fixed HeLa cells expressing Lifeact pep0-pep9 in the absence of SZ21. Total cellular actin was visualized by Phalloidin staining (magenta). Lifeact pepCAST (pep6) was selected for further study since its localization to F-actin was significantly reduced (yellow arrows). Scale bar, 30 µm. d Quantification of F-actin binding inhibition for Lifeact pep0-pep9, where 0% is defined by Lifeact-only localization and 100% is defined by GFP-only localization. Bars represent mean ± SD. n = 40 biological replicates. e Fluorescent micrographs of pepCASTs, SZ21, and the combination of pepCASTs and SZ21 in live HeLa cells (left). After SZ21 expression, pepCAST localizes to F-actin filaments. Quantification of binding activation (right), where 0% is defined by inactive pepCAST localization and 100% is defined by ABM-only localization. Scale bar, 30 µm. Bars represent mean ± SD. n = 20 biological replicates. f Time-lapse imaging of pepCASTs shows increased localization of pepCASTs to F-actin over time (white arrows). SZ21 expression was induced with 1 µM ATc to stable pepCAST-expressing HeLa cells transfected with SZ21 in an ATc-inducible plasmid. Scale bar, 30 µm. g Kinetics of pepCASTs’ activation. The ratio of pepCAST’s F-actin-localized fluorescence intensity to its cytoplasmic fluorescence intensity normalized to GFP-only localization is plotted at several timepoints, up to 7 h. t1/2 = binding half-time. n = 3 biological replicates. Data were fit to a one-phase exponential association model (broken lines). P values were determined using a two-tailed unpaired t-test comparison with ABM-only control (b, d) or inactive pepCAST (e). (ns not significant P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001). Source data are provided as a Source Data file.
With pepCAST candidates, we turned our attention to the activation of the ABM upon the introduction of SZ21. The presence of SZ21 should disrupt SZ3-SZ4 intramolecular binding and enable ABM engagement with F-actin. To test this, we expressed pepCASTs of ABS and Lifeact in the presence or absence of SZ21. Only in the presence of SZ21 did activation and F-actin engagement of pepCASTs occur (Fig. 3e), with the ABS pepCAST reaching >80% activation. The stimulus alone, SZ21, was entirely cytoplasmic, but upon dual expression with pepCASTs, SZ21 transitioned to decorating F-actin filaments, consistent with an intermolecular interaction leading to activation of the pepCASTs. We did not observe a correlation between single-cell CAST expression level and activation (Supplementary Fig. 4), suggesting that the concentration of actin far exceeds the concentration of pepCASTs, giving rise to expression-independent activation. As well, we verified pepCAST activation in vitro with purified forms of pepCAST and F-actin (Supplementary Figs. 5a, 6a, b) and confirmed that pepCASTs do not alter actin polymerization in vitro (Supplementary Fig. 5b).
To determine the kinetics of pepCAST activation, we first generated stable cells expressing either the ABS pepCAST or the Lifeact pepCAST. With these cells, we expressed SZ21 from an inducible promoter (Fig. 3f). SZ21 expression was observable 2 h after induction with Anhydrotetracycline (ATc) (Supplementary Fig. 3), and we quantified pepCAST F-actin engagement from this time point over the course of 7 h. Both pepCASTs activated as pseudo first-order rate processes on the timescale (t1/2) of 2–3 hs (Fig. 3g and Supplementary Movie 1). Our results suggest that intramolecular binding, indeed, leads to inactivity of both ZO-1’s ABS and Lifeact, and that F-actin engagement can be restored by disrupting intramolecular binding with a stimulus, in this case SZ21.
Small molecule-based CAST leads to faster activation
Encouraged by our pepCAST results, we next sought to develop other CAST systems with different turn-on kinetics and stimuli, providing tools that can be used for a variety of actin experiments requiring compatible turn-on times and involving multiple actin-binding proteins. Small molecules as stimuli offer fast turn-on kinetics since they can readily diffuse through the cell membrane and bind to their targets with large rate constants46,47. Several small molecules are known to bind tightly to both the catalytically competent and the catalytically dead NS3a protease from the hepatitis C virus and have been used previously to trigger cellular processes48,49,50,51. To take advantage of these small molecules as stimuli for CASTs, we constructed several possible CAST constructs by fusing catalytically dead NS3a and an NS3a binding ligand (CP5-46A-4D5E, referred to here as CP5)52, to the termini of the ABMs with and without linkers (Fig. 4a). One arrangement, sm2, caused noticeable disruption to F-actin engagement (Fig. 4b–d). Using AlphaFold253,54, we compared a model for the ABS to that of sm2, which we termed smCAST (Fig. 4c and Supplementary Fig. 7). The model indicated that ABS in isolation sits as an α-helix. However, within smCAST, the ABS appeared constrained, where it adopts a disordered structure, a conformational change that would inhibit ABS’s F-actin activity when embedded in smCAST.
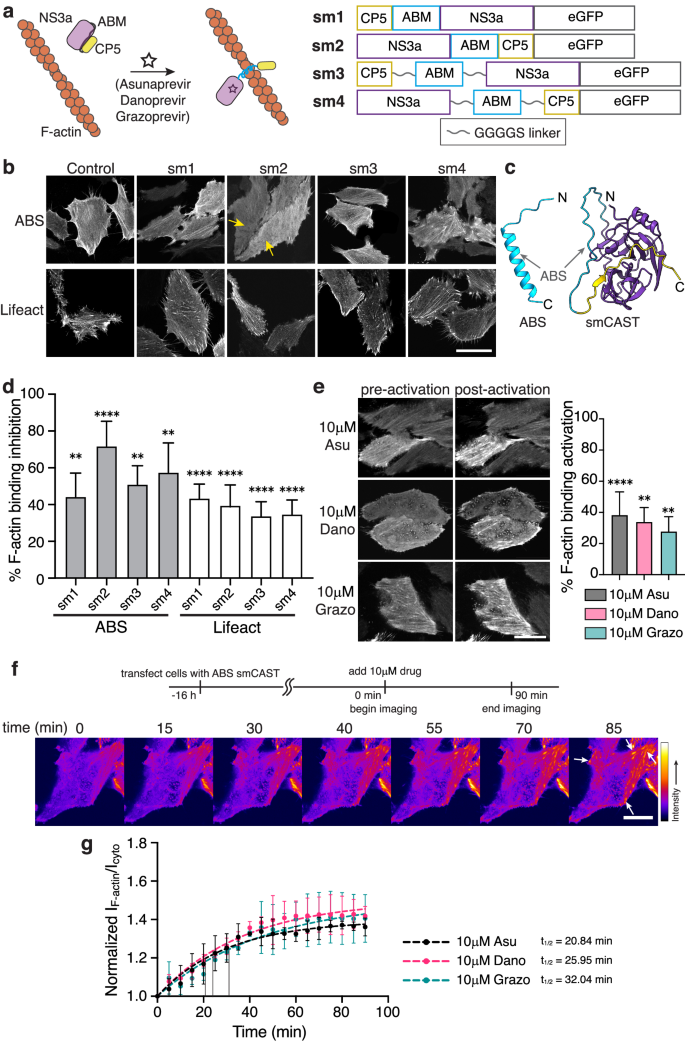
a Schematic of a small molecule-based CAST system. The ABM is initially constrained (gray) by the intramolecular association of NS3a and CP5. After the addition of small molecule inhibitors (Asunaprevir, Danoprevir, or Grazoprevir) that disrupt the NS3a:CP5 complex, the CAST transitions to an active state (blue) capable of F-actin binding (left). The primary sequence of small molecule-based CAST candidates tested (right). b Fluorescent micrographs of fixed HeLa cells expressing sm1-sm4 in the absence of small molecule inhibitors. smCAST (sm2) was selected for further study since its localization to F-actin was significantly reduced (yellow arrows). Scale bar 30 µm. c AlphaFold2 prediction of native ABS (blue, left) and smCAST (right). ABS alone is predicted to be an α-helix, the active form of the ABM, whereas within smCAST the ABS sequence is unstructured (right). d Quantification of F-actin binding inhibition for ABS and Lifeact sm1-sm4, where 0% is defined by ABS-only localization and 100% is defined by GFP-only localization. Bars represent mean ± SD. n = 40 biological replicates. e Fluorescent micrographs of smCAST in the presence or absence of Asu, Dano, or Grazo in live HeLa cells (left). Quantification of activation in the presence of drug for 30 min (right), where 0% is defined by inactive smCAST localization and 100% is defined by ABS-only localization. Scale bar 20 µm. Bars represent mean ± SD. n = 30 biological replicates. f Time-lapse imaging of the smCAST after the addition of Grazo shows increased localization of smCAST to F-actin (white arrows) over time. Scale bar 10 µm. g Kinetics of smCAST activation. The ratio of smCAST’s F-actin-localized fluorescence intensity to its cytoplasmic fluorescence intensity normalized to GFP-only localization is plotted at several timepoints, up to 90 min. t1/2 = binding half-time. n = 3 biological replicates. Data were fit to a one-phase exponential association model (broken lines). P values were determined using a two-tailed unpaired t-test comparison with ABS-only control (d) or inactive smCAST (e). (ns not significant P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001). Source data are provided as a Source Data file.
To examine the activation of smCAST, we incubated smCAST-expressing HeLa cells with either Asunaprevir (Asu), Danoprevir (Dano), or Grazoprevir (Grazo), small molecules known to bind tightly to NS3a’s active site55,56. In all cases, activation of smCAST and corresponding F-actin engagement occurred, with Asu giving rise to the highest level of activation (Fig. 4e). We verified activation of smCAST in vitro upon the addition of small molecule stimulus (Supplementary Fig. 6c) and confirmed that smCAST does not alter actin polymerization in vitro (Supplementary Fig. 5b). In cells, the extent of smCAST activation was influenced by the concentration of the small molecule stimulus, suggesting a dose-dependent smCAST response (Supplementary Fig. 8). To confirm that activation was due to small molecule binding to NS3a, we cloned two NS3a mutants57, D182V and D182Y, that do not recognize Asu, in place of NS3a in smCAST. Unlike smCAST, the two variants did not re-localize to F-actin in response to 10 µM Asu (Supplementary Fig. 9), indicating that activation of smCAST is due to direct binding of small molecule stimuli.
To examine smCAST kinetics, we visualized smCAST over time in cells that were treated with different drugs. Similar to pepCASTs, smCAST followed pseudo first-order turn-on kinetics. However, in contrast to pepCASTs, smCAST activated faster than pepCASTs, on the timescale (t1/2) of 20 to 30 min, with Asu providing the fastest response of the small molecules tested (Fig. 4f, g and Supplementary Movie 2). This data suggests that varying the intramolecular binders and types of stimuli can lead to additional CAST systems with different turn-on kinetics.
Rapid activation of light-based CASTs
pepCAST and smCAST activate on hours and tens of minutes timescales, respectively. Still, some cellular processes occur on shorter timescales (single minutes) and, as such, demand more rapid turn-on58,59. Light-sensitive protein domains, for example, LOV60,61 and CRY62,63 domains, are known to isomerize and conformationally change on shorter timescales64 than pepCAST’s and smCAST’s activation. To test whether light can act as a stimulus for CAST activation, we designed optical CASTs, opto1-opto2, by fusing either AsLOV2-Ja’ or a mutant with stronger intramolecular association (AsLOV2 L514J-Ja’ L531E)65 to the ABMs. LOV2 domains can sterically occlude short peptide sequences65,66, which motivated us to examine protein architectures with ABMs at the terminus. For both opto1 and opto2, we observed significant inhibition of F-actin-binding activity, with Lifeact opto1 reaching ~90% inhibition (Fig. 5b, c). The opto1 constructs, termed optoCASTs, were subsequently tested for activation with blue light illumination. This resulted in a rapid and effective redistribution of optoCASTs to F-actin in cells (Fig. 5d), achieving 80% activation, with a t1/2 of 3–5 min (Fig. 5e, f and Supplementary Movie 3). Localized activation in individual cells was also possible with optoCAST (Supplementary Fig. 10), showcasing one advantage to a light stimulus. As with pepCASTs and smCAST, purified forms of ABS and Lifeact optoCASTs (Supplementary Fig. 5a) activated in response to blue light illumination in vitro (Supplementary Fig. 6d, e), and the presence of activated optoCASTs did not alter actin polymerization (Supplementary Fig. 5b).
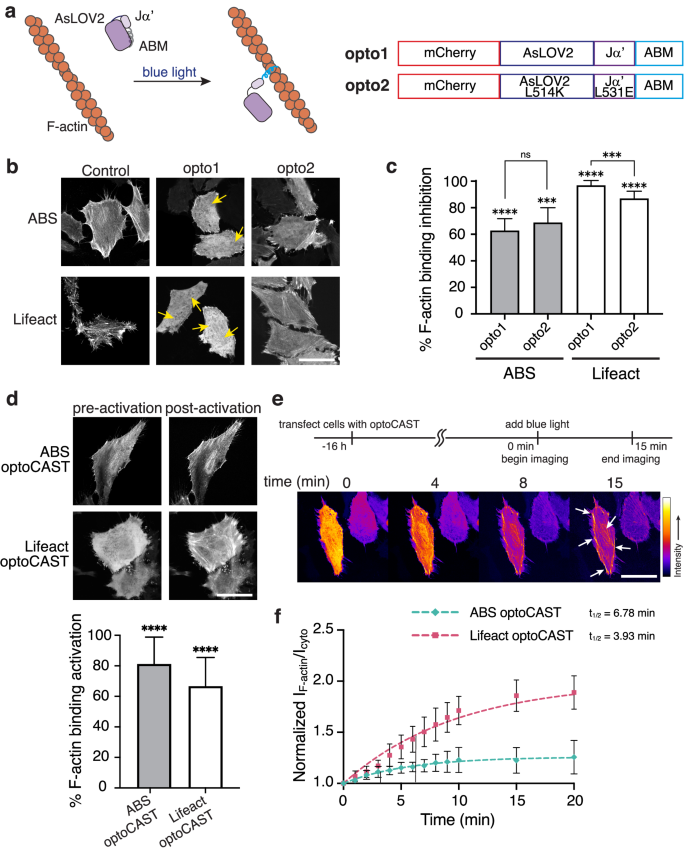
a Schematic of a light-based CAST system. The ABM is initially occluded (gray) by the AsLOV2 domain until blue light illumination leads to the active state (blue) capable of F-actin binding (left). The primary sequence of light-based CAST candidates tested (right). b Fluorescent micrographs of fixed HeLa cells expressing opto1-opto2 in the absence of blue light. ABS and Lifeact optoCASTs (opto1) were selected for further study since their localization to F-actin was significantly reduced (yellow arrows). Scale bar 30 µm. c Quantification of F-actin binding inhibition for ABS and Lifeact opto1-opto2, where 0% is defined by ABM-only localization and 100% is defined by mCherry-only localization. Bars represent mean ± SD. n = 20 biological replicates. d Fluorescent micrographs of optoCASTs before and after blue light illumination in live HeLa cells (top). Quantification of activation (bottom), where 0% is defined by inactive optoCAST localization and 100% is defined by ABM-only localization. Scale bar 20 µm. Bars represent mean ± SD. n = 20 biological replicates. e Time-lapse imaging of Lifeact optoCAST under blue light illumination shows increased localization of optoCAST to F-actin (white arrows) over time. Scale bar 20 µm. f Kinetics of ABS and Lifeact optoCAST activation. The ratio of optoCAST’s F-actin-localized fluorescence intensity to its cytoplasmic fluorescence intensity normalized to mCherry-only localization is plotted at several timepoints, up to 20 min. t1/2 = binding half-time. n = 3 biological replicates. Data were fit to a one-phase exponential association model (broken lines). P values were determined using a two-tailed unpaired t-test comparison with ABM-only control (c) or inactive optoCAST (d). (ns not significant P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001). Source data are provided as a Source Data file.
With smCAST and optoCAST—two switches that can be activated within the same order of magnitude—in hand, we next examined whether orthogonal triggering was possible. To do this, we co-expressed smCAST and Lifeact optoCAST in cells and triggered activation with either small molecules, such as Asu, Dano, and Grazo, or blue light. In the presence of small molecule stimuli, activation of smCAST ensued with the re-localization of smCAST to F-actin, while optoCAST remained cytoplasmic. Similarly, blue light stimulus caused optoCAST to decorate F-actin, whereas smCAST remained cytoplasmic. Sequential activation within the same cell was also possible (Supplementary Fig. 11). Together, these experiments show that each CAST is sensitive to its distinct stimulus and that multiple CASTs can be used simultaneously for multiplexed and orthogonal control over F-actin binding in cells.
Dimeric optoCASTs enable control over F-actin architecture and cell shape with light
By varying F-actin’s organization, cells regulate their shape, movement, and mechanics. Bundled filaments, for instance, represent a prominent F-actin architecture in stress fibers and at the cell cortex13,67,68. CASTs offer an opportunity to induce F-actin re-organization and subsequent cellular responses by engineering the intramolecular-bound states into structures that can alter F-actin architecture dynamically. Dimeric ABD-containing proteins, so-called actin crosslinkers, are responsible for forming bundled filaments in the cell69,70,71, and we reasoned that generating a dimeric CAST should provide control over filament bundling in a user-defined manner. To construct multimeric CASTs in cells, we appended a self-dimerizing peptide sequence to the N-terminus of optoCASTs, termed dOptoABS and dOptoLifeact (Fig. 6a), and expressed these sequences in HEK 293 cells, which lack extensive bundled filaments (Supplementary Fig. 12a). After illuminating with blue light, we observed dramatic re-organization of F-actin into heavily crosslinked architectures at both the cell periphery and in the cell body for both dOptoABS and dOptoLifeact (Fig. 6b and Supplementary Movie 4). Coincident with crosslinking, cells also appeared to contract after blue light triggering. Quantification of cell area indicated an area reduction of approximately 10% for dOptoABS and 20% for dOptoLifeact upon activation. Next, we wondered whether triggering actin crosslinking and cell area changes would be possible in more crowded environments, such as in tissues. We, consequently, expressed either dOptoABS or dOptoLifeact in MDCK II epithelial monolayers and activated with blue light. After blue light illumination, we observed similar F-actin re-organization as in isolated cell experiments (Fig. 6c and Supplementary Movie 5), but, in the case of tissue, we also found that cell contacts were disrupted with the majority of cells becoming detached from their neighbors after dimeric CAST activation. These experiments show that CASTs are amenable to further engineering for synthetic biology applications, where control over F-actin organization is desired.
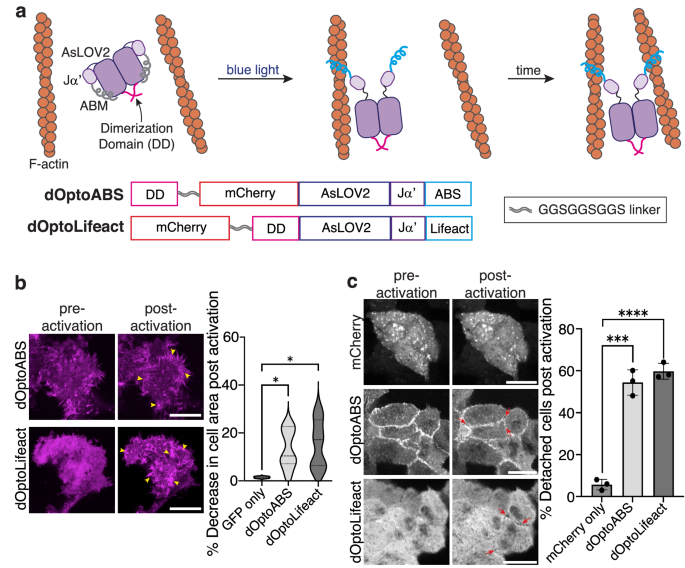
a Schematic of the design and activation of a dimeric optoCAST. A self-associating peptide (pink) is tethered to either ABS optoCAST (dOptoABS) or Lifeact optoCAST (dOptoLifeact) to form dimeric versions of optoCAST. Photoactivation of the dimeric species would then expose the two occluded ABMs, leading to actin crosslinking and bundling in cells (top). Primary sequences of dOptoABS and dOptoLifeact (bottom). b Fluorescent micrographs of HEK 293 T cells expressing either dOptoABS or dOptoLifeact before and after blue light illumination (left). Clusters of bundled filaments are present post-activation (yellow arrowhead). Quantification of cell area changes following activation (right). Scale bar 20 µm. n = 12 biological replicates. c Fluorescent micrographs of MDCK II cell islands expressing either dOptoABS or dOptoLifeact before and after blue light illumination (left). Cells contract within the island and detach from one another (red arrows). Quantification shows an increase in cell detachment post-activation (right). Scale bar 15 µm. Bars represent mean ± SD. n = 25 cell-cell contacts. P values were determined using a two-tailed unpaired t-test comparison with GFP-only control (b) or mCherry-only control (c). (ns not significant P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001). Source data are provided as a Source Data file.
Manipulating proteins and cellular processes dynamically by replacing native ABDs with CASTs
Finally, we sought to test whether CASTs can be used in place of a native ABD to control protein activity in cells. To do this, we focused on the junctional protein, ZO-1, which plays an active role in focal adhesion formation and cell migration72,73. ZO-1, a major component of the tight junction in epithelial tissue, also helps drive persistent migration of subconfluent cells by interacting with integrin α5β1 and regulating focal adhesions74,75. We expressed either wildtype ZO-1 (WT ZO-1) or ZO-1 lacking its ABS (ZO-1ΔABS) in subconfluent cells (Fig. 7a) and found that, indeed, WT ZO-1 localizes to focal complexes at the cell periphery and to larger focal adhesion clusters under the cell body (Fig. 7b). In contrast, ZO-1ΔABS primarily localized to small focal complexes on the cell periphery, indicating that ZO-1’s engagement with F-actin modulates its adhesion activity. Next, we constructed a version of ZO-1 that replaces ZO-1’s native ABS sequence with our ABS smCAST, termed ZO-1smCAST (Fig. 7a). From our data above, we anticipated that if actin-binding of ZO-1 occurred, then the protein would populate larger focal adhesion clusters over time. By fluorescence microscopy, we first imaged ZO-1smCAST and found that it localizes to small focal complexes at the cell periphery (Fig. 7c), similar to ZO-1ΔABS. After activation with either Asu, Dano, or Grazo, however, ZO-1smCAST re-localized over the course of 60 min, occupying larger focal adhesions under the cell body and mirroring that of WT ZO-1 (Fig. 7c and Supplementary Movie 6), (Fig. 7d, e). Encouraged by these results, we then turned to a wound healing assay of collective cell migration. We’ve previously shown that ZO proteins, including ZO-1, are essential for epithelial cell migration using a knockout line for ZO-1 and ZO-273. To this knockout line, we expressed either WT ZO-1 or ZO-1smCAST and performed a wound healing experiment. WT ZO-1-expressing cells were able to migrate and close the wound area over the course of 24 h, whereas the ZO-1smCAST-expressing cells did not close the wound area after 52 h (Fig. 7f–h), suggesting that ZO-1’s ability to bind to F-actin is necessary for collective migration. To test this directly, we repeated the wound healing assay with ZO-1smCAST-expressing cells and added Asu, Dano, or Grazo to activate the smCAST module within ZO-1’s structure. Gratifyingly, in all cases, ZO-1smCAST-expressing cells were able to migrate and close the wound area in the presence of small molecule stimuli on timescales similar to WT ZO-1-expressing cells (Fig. 7f–i and Supplementary Fig. 13a–c). Small molecule stimuli, Asu, Dano, or Grazo, had no effect on WT ZO-1-expressing cells. Taken together, our data show that CASTs can be incorporated into the native sequence of proteins to control their engagement with F-actin in both cells and tissue.
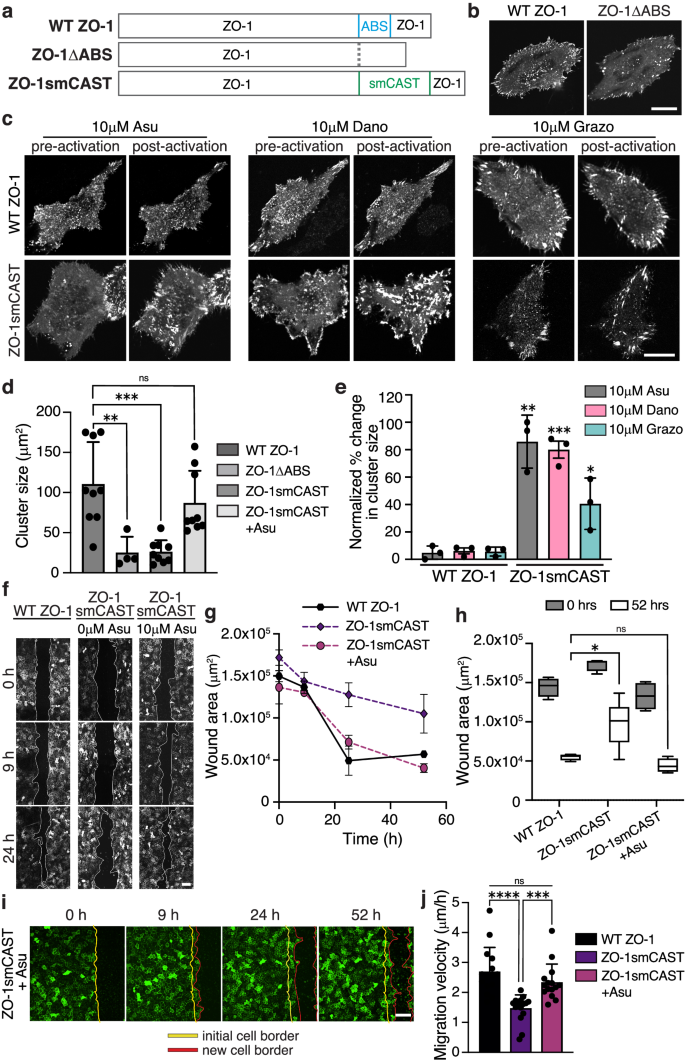
a Primary sequence of full-length WT ZO-1, ZO-1 with its ABS deleted (ZO-1ΔABS), and ZO-1 with its ABS replaced with smCAST (ZO-1smCAST). b Fluorescent micrographs of WT ZO-1 and ZO-1ΔABS expressed in live HeLa cells. ZO-1 localizes to focal complexes and large clusters of focal adhesions under the cell body, while ZO-1ΔABS localizes primarily to peripheral focal complexes. Scale bar 20 µm. c Fluorescent micrographs of WT ZO-1 and ZO−1smCAST before and after Asu, Dano, or Grazo addition in live HeLa cells. After activation, only ZO−1smCAST re-localizes to large focal adhesion clusters (lower panel). Scale bar 20 µm. d Quantification of ZO-1 cluster size in HeLa cells expressing ZO-1 variants. Bars represent the mean ± SD of clusters. n = 3 biological replicates. e Change in cluster size of ZO-1 variants before and after drug treatment. Bars represent mean ± SD. n = 3 biological replicates. f Fluorescent micrographs of collective cell migration in a wounded MDCK II monolayer over time. Stable cells lacking ZO proteins and expressing either WT ZO-1 or ZO-1smCAST were imaged with and without Asu. Scale bar 100 µm. g Quantification of wound area over time for cells expressing WT ZO-1 or ZO-1smCAST and cultured in the absence or presence of Asu. Data were presented as mean ± SD. n = 3 biological replicates. h Quantification of wound area after 52 h. The box represents 25th to 75th percentiles with the middle line as the median and whiskers as the maximum and minimum values. n = 4 biological replicates. i Time-lapse imaging of wound healing for ZO-1smCAST-expressing MDCK II cells treated with Asu. A leading edge of cells (red line) migrates from an initial wound boundary (yellow line) during wound healing. Scale bar 100 µm. j Quantification of the migration rate of cells expressing ZO-1 variants. Bars represent mean ± SD. n = 15 biological replicates. P values were determined using a two-tailed unpaired t-test. Comparison is to WT ZO-1-expressing cells treated with the same drugs (e). (ns not significant P > 0.05; *P < 0.05; **P < 0.01; ****P< 0.0001). Source data are provided as a Source Data file.
Concluding remarks
F-actin plays numerous roles throughout the cell, and a diverse set of proteins bearing ABDs orchestrate the varied activities of F-actin. Identifying which ABD-containing proteins are necessary for certain cellular processes and defining their time evolution of actin engagement remains challenging due to a lack of compatible probes and molecular tools. Here, we developed CASTs, which enable control over ABD activity in response to peptide, small molecule, or light stimuli. CASTs add to a growing toolbox of engineered proteins that can be expressed and subsequently activated to probe the influence of F-actin76,77,78,79,80. Being protein-based, these tools offer notable advantages since they are genetically encoded and can be expressed in cells that may be difficult to treat with exogenous probes. Most of the previous tools, however, exert their activity on all actin filaments, either by perturbing polymerization or depolymerization of actin76,78 or by severing of F-actin77,79, in turn impacting numerous ABDs. CASTs differ by controlling the binding of a single ABD to F-actin, keeping the actin filament and the other bound ABDs intact and engaged.
The suite of CASTs provides different timescales of activation and the possibility of multiplexing as each CAST module can be triggered orthogonally. Further, orthogonal activation allows control of actin-binding at distinct subcellular locations at different times and in varied order, all within the same cell. With multiple options for activation, CASTs can accommodate a wide variety of experimental demands. Activation deep within organisms can often be challenging with exogenous reagents or light, yet pepCASTs can be activated by a genetically encoded peptide stimulus, enabling control of actin-binding in vivo. optoCAST, on the other hand, is well suited for sequential activation of individual cells during a development process or in subcellular locations in culture. For control over the single and tens of minutes timescale of activation, optoCAST and smCAST will be the most useful, while long timescale, sustained activation will be facile with pepCAST. We showed that CASTs can be inserted into native protein sequences, allowing user-defined manipulation of single protein ABD activity in a background of abundant and diverse ABDs. For this use of CASTs, the target ABD-containing protein must be amenable to protein fusions. We also found that CASTs can be further engineered into synthetic structures, creating architectures that re-organize the cytoskeleton. CASTs present a much-needed strategy for uncovering key aspects of the cytoskeleton and for manipulating cell mechanics and behavior in a user-defined manner.